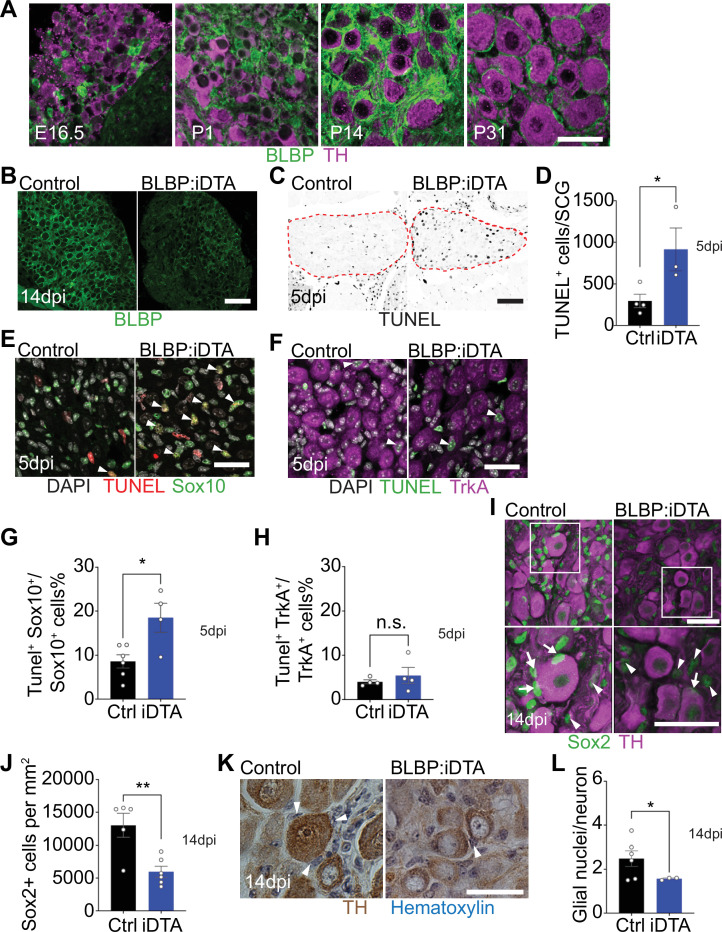
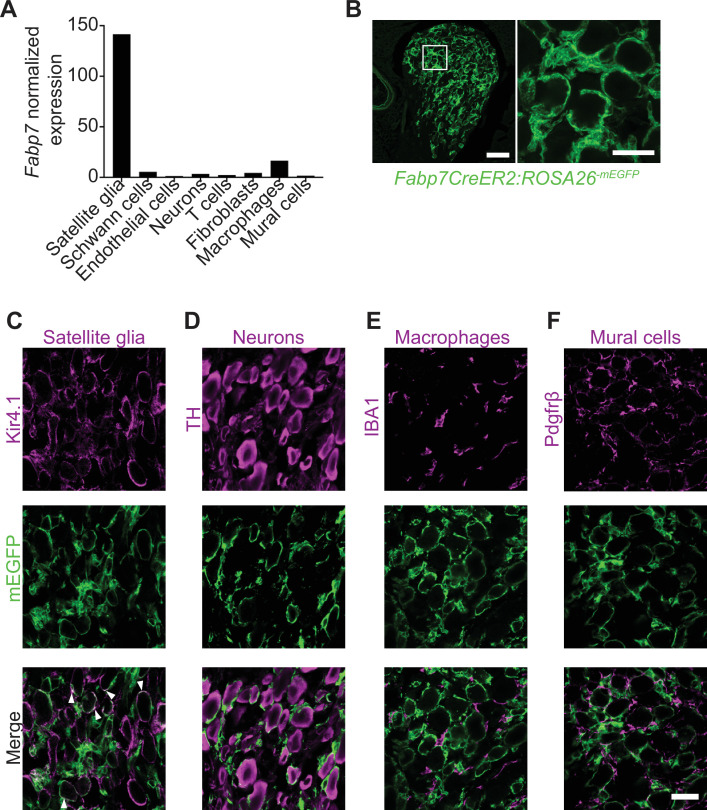
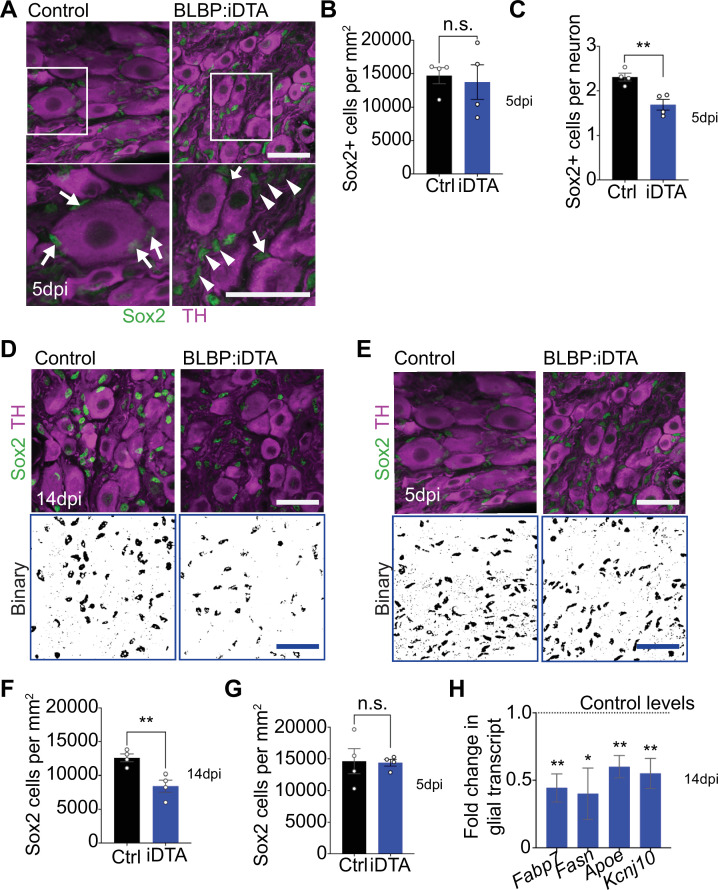
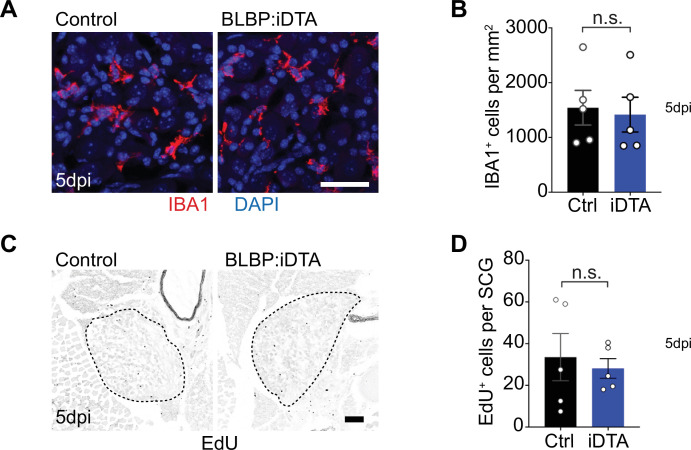
Figure 1. DTA-mediated ablation of satellite glia in sympathetic ganglia.
(A) Satellite glial cells, immunolabeled with brain lipid binding protein (BLBP, green), progressively ensheathe sympathetic neuron cell bodies, labeled with tyrosine hydroxylase (TH, magenta) in the superior cervical ganglia (SCG) during development. Time points shown are embryonic day 16.5 (E16.5) and postnatal days P1, P14, and P31. Scale bar: 30 μm. (B) Reduced BLBP expression in BLBP:iDTA SCG relative to control ganglia at 14 days after the last tamoxifen injection (14 dpi). Scale bar: 100 μm. (C) Increased apoptosis in BLBP:iDTA SCG (outlined in red dashed line) compared to controls as detected by TUNEL labeling at 5 days post-tamoxifen injection (5 dpi). Scale bar: 100 μm. (D) Quantification of apoptotic cells in SCGs at 5 dpi from n = 4 control and 3 mutant mice. Data are means ± SEM. *p<0.05, t-test. (E) Increased apoptosis of satellite glial cells assessed by Sox10 immunostaining (green) and TUNEL labeling (red) in BLBP:iDTA sympathetic ganglia compared to control at 5 dpi. DAPI channel is shown in gray. Arrowheads indicate TUNEL+;Sox10+ cells. Scale bar: 30 µm. (F) Neuronal apoptosis is similar between BLBP:iDTA and control ganglia at 5 dpi as assessed by TrkA immunostaining (magenta) and TUNEL labeling (green). DAPI channel is shown in gray. Arrowheads indicate TUNEL+;TrkA+ neurons. Scale bar: 30 µm. (G) Quantification of TUNEL+;Sox10+ cells expressed as a % of Sox10+ cells. Data are presented as means ± SEM from n = 6 control and 4 mutant mice, **p<0.05, t-test. (H) Quantification of TUNEL+;TrkA+ cells expressed as a % of TrkA+ cells. Data are presented as means ± SEM from n = 4 mice per genotype, n.s, not significant, t-test. (I) Decreased association of Sox2-labeled satellite glia (green) with TH-positive sympathetic neurons (magenta) in BLBP:iDTA sympathetic ganglia compared to controls at 14 dpi. Arrows indicate Sox2-labeled nuclei of satellite glia associated with TH-positive sympathetic neuron cell bodies, while arrowheads indicate Sox2-labeled nuclei not abutting neuronal soma. Gain in both images has been set to the same level for a better visualization of Sox2-labeled nuclei in mutant ganglia. Scale bar: 30 μm. (J) Quantification of Sox2-positive cells in control and BLBP:iDTA SCGs at 14 dpi. Data are presented as mean ± SEM from n = 5 control and 6 mutant animals, **p<0.01, t-test. (K) TH DAB immunohistochemistry and hematoxylin staining shows fewer satellite glia nuclei (blue) associated with TH-labeled sympathetic neuron cell bodies (brown) in BLBP:iDTA ganglia at 14 dpi. Scale bar: 30 µm. (L) Quantification of glial nuclei associated with neuronal soma. Data are presented as means ± SEM from n = 6 control and 3 mutant animals, **p<0.05, t-test.