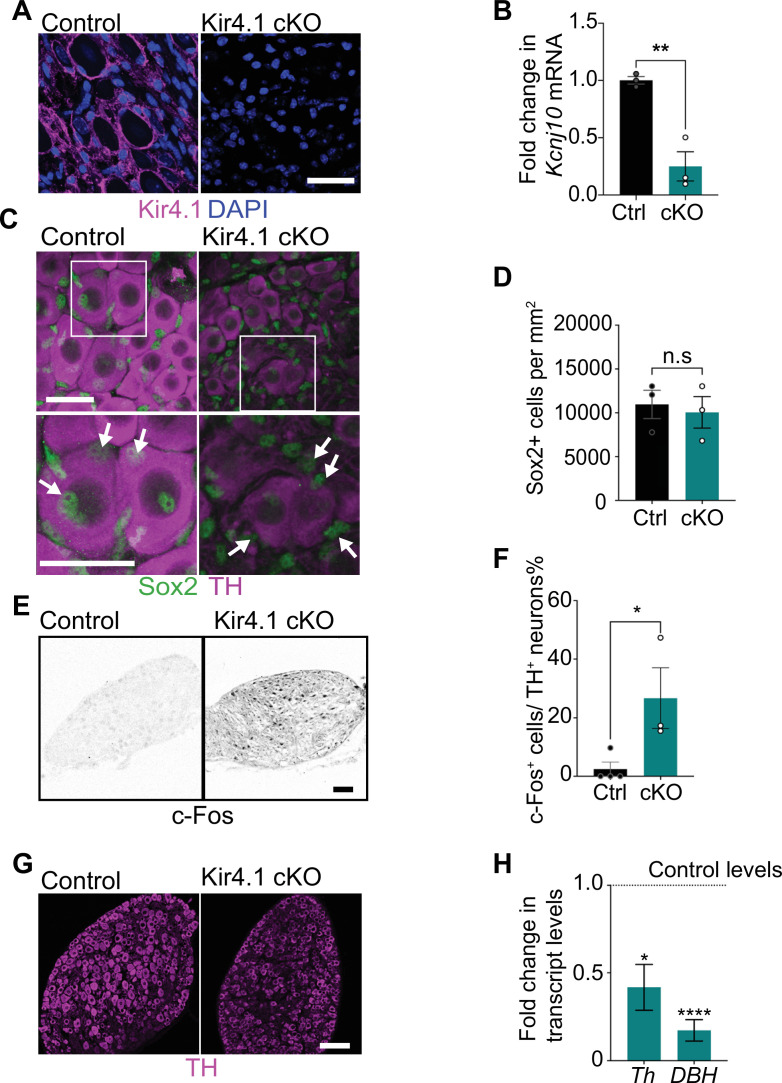
Figure 4. Satellite glia-specific Kir4.1 loss impairs norepinephrine (NE) enzyme expression and neuron activity.
(A, B) Reduced Kir4.1 protein and transcript (Kcnj10) in Kir4.1 cKO mice. Scale bar: 30 μm. Data in (B) are mean ± SEM from n = 3 animals per genotype, **p<0.01, t-test. (C, D) Sox2-positive satellite glial cell numbers are unaffected in Kir4.1 cKO SCG. Arrows indicate Sox2-labeled nuclei of satellite glia associated with sympathetic neuron cell bodies. Arrowheads indicate Sox2-labeled satellite glia that are not adjacent to neuronal soma. Scale bar: 30 μm. Data are presented as mean ± SEM from n = 3 animals per genotype, n.s., not significant, t-test. (E, F) Kir4.1 deletion in satellite glia results in an increase in c-Fos-positive sympathetic neurons. Quantification of c-Fos+;TH+ sympathetic neurons as a % of total number of TH+ sympathetic neurons. Scale bar: 100 μm. Quantifications are mean ± SEM from n = 3 animals per genotype, *p<0.05, t-test. (G, H) Downregulation of NE biosynthetic enzymes, TH and DBH, in Kir4.1 cKO sympathetic ganglia. Results are mean ± SEM from n = 3–5 animals per genotype, *p<0.05, ****p<0.0001, t-test with Bonferroni’s correction.