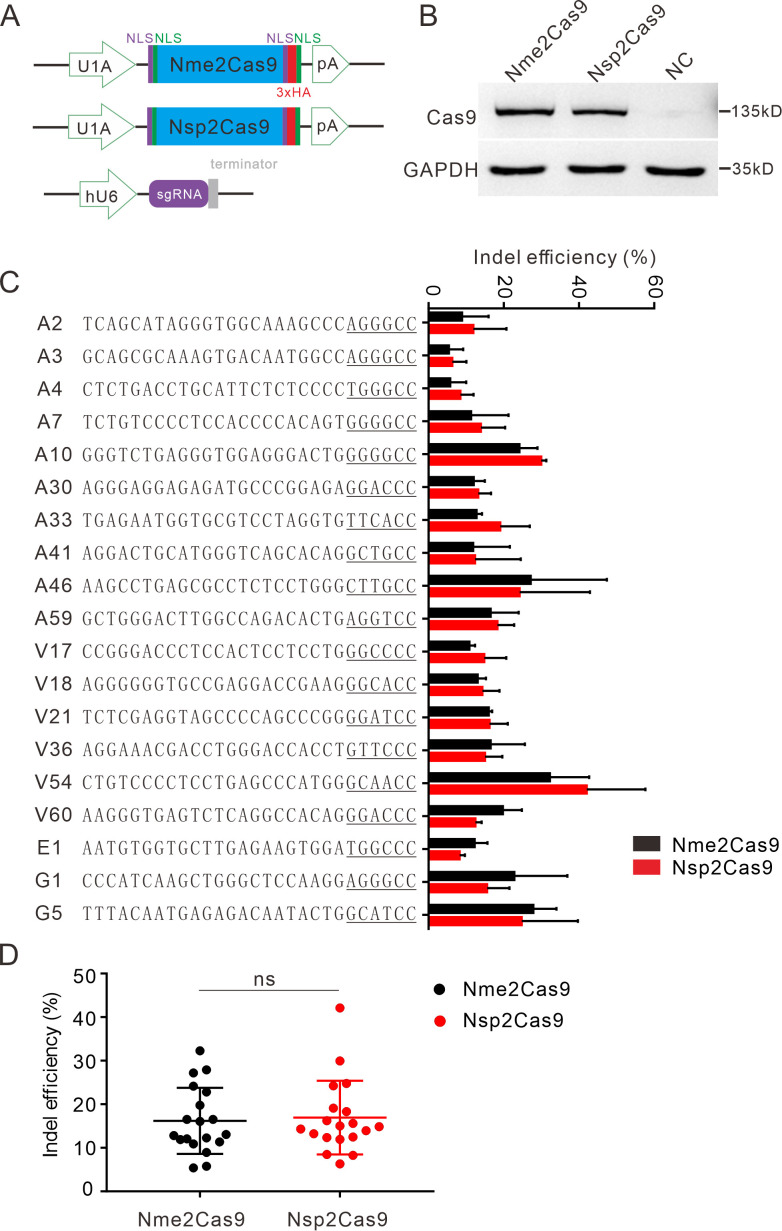
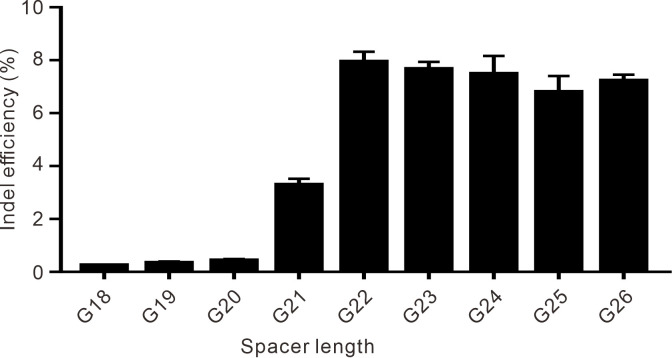
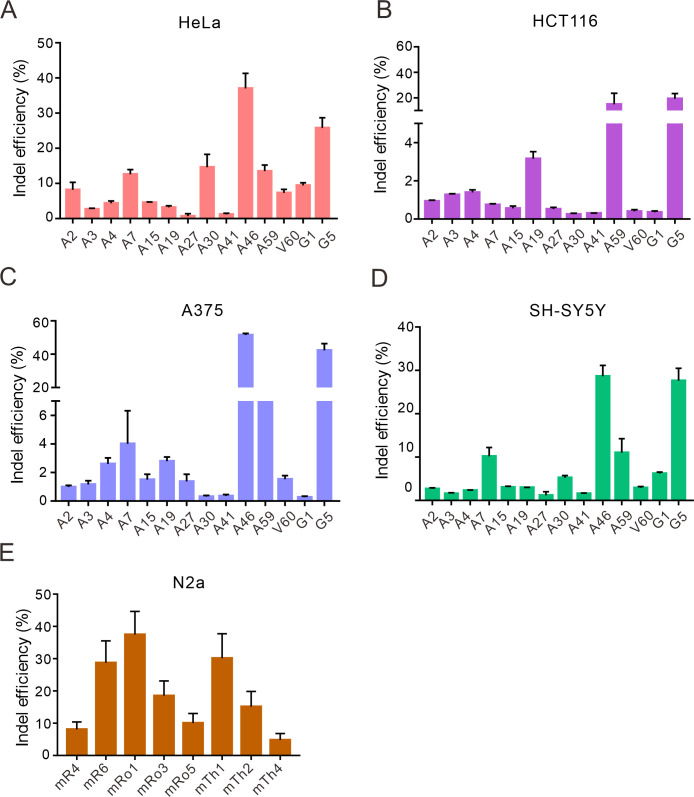
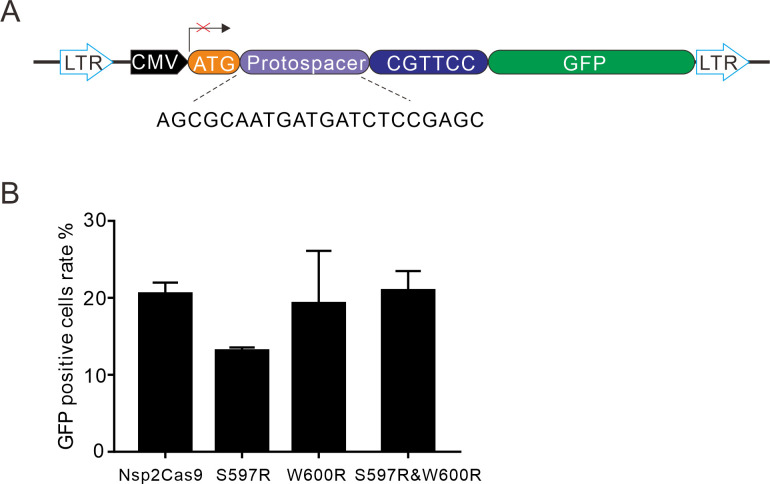
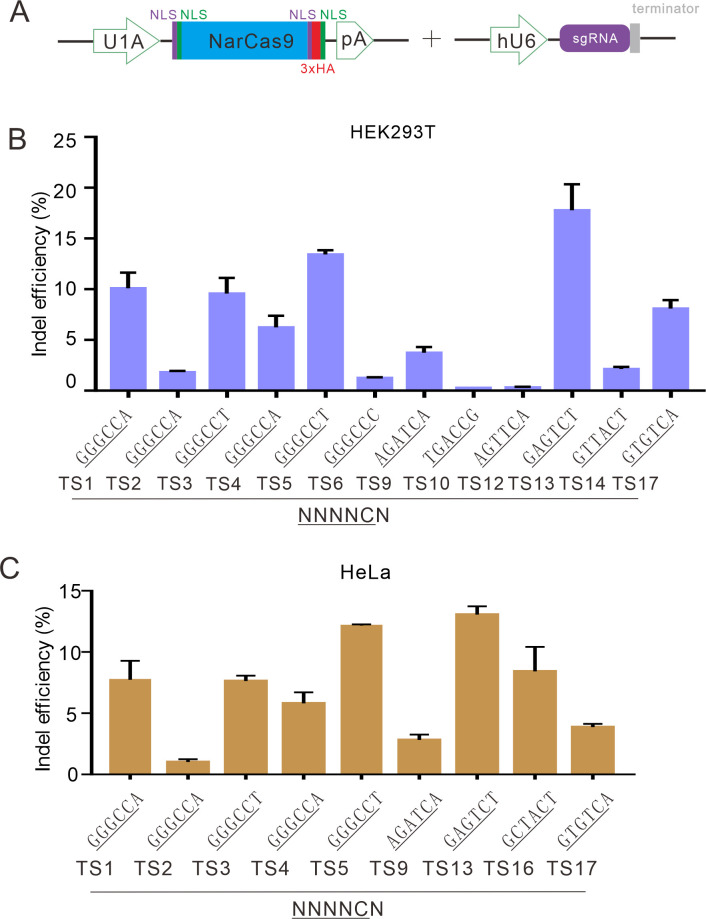
Figure 3. Nsp2Cas9 enables editing in HEK293T cells.
(A) Schematic of Cas9 and sgRNA expression constructs. U1A: U1A promoter; pA: polyA; NLS: nuclear localization signal; HA: HA tag. (B) Protein expression levels of Nsp2Cas9 and Nme2Cas9 were analyzed by Western blot. HEK293T cells without Cas9 transfection were used as negative control (NC). (C) Comparison of Nsp2Cas9 and Nme2Cas9 editing efficiencies at 19 endogenous loci in HEK293T cells. Data represent mean ± SD for n=3 biologically independent experiments. (D) Quantification of the indel efficiencies for Nsp2Cas9 and Nme2Cas9. Each dot represents an average efficiency for an individual locus. Data represent mean ± SD for n=3 biologically independent experiments. P values were determined using a two-sided Student’s t test. P=0.7486 (P>0.05), ns stands for not significant.