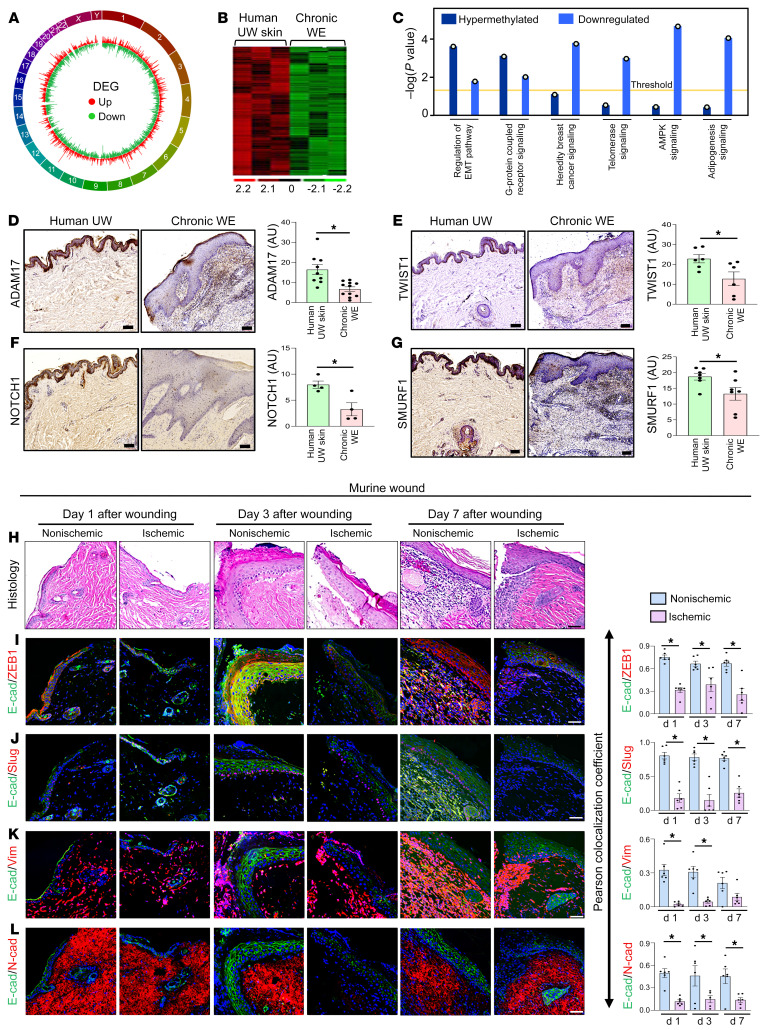
Figure 2. Increase in global DNA methylation represses EMT pathway in chronic WE tissue.
(A) Circos plot demonstrating the distribution of significant differentially expressed genes (DEGs) obtained through RNA-Seq analysis in chronic WE. Chromosome number marked in the periphery. Red lines represent upregulated genes and green lines represent downregulated genes in chronic WE. (B) Hierarchical clustering analysis of significantly different genes in chronic WE. (C) Bar plot representing integration of hypermethylated genes (by MethylCap-Seq analysis) with downregulated genes (by RNA-Seq analysis) was performed to look for the common canonical pathways using the comparison analysis tool of IPA. The dot over each bar represents the –log(P value) of each pathway. (D) Representative IHC analysis of ADAM17, (E) TWIST1, (F) NOTCH1, and (G) SMURF1 in paraffin sections of human UW skin and chronic WE. Right panels represent the intensity analysis of the images. (Scale bar: 100 μm; *P < 0.05, Student’s t test, n = 4–11). (H) H&E staining and (I) representative IHC analyses for E-cadherin–ZEB1 colocalization, (J) E-cadherin–slug colocalization, (K) E-cadherin–vimentin colocalization, and (L) E-cadherin–N-cadherin colocalization in ischemic and nonischemic murine bipedicle wounds at different time points after wounding. Right panels represent the Pearson colocalization coefficient calculation at days 1, 3, and 7 after wounding. (Scale bar: 50 μm; n = 6, *P < 0.05). Data represented as mean ± SEM.