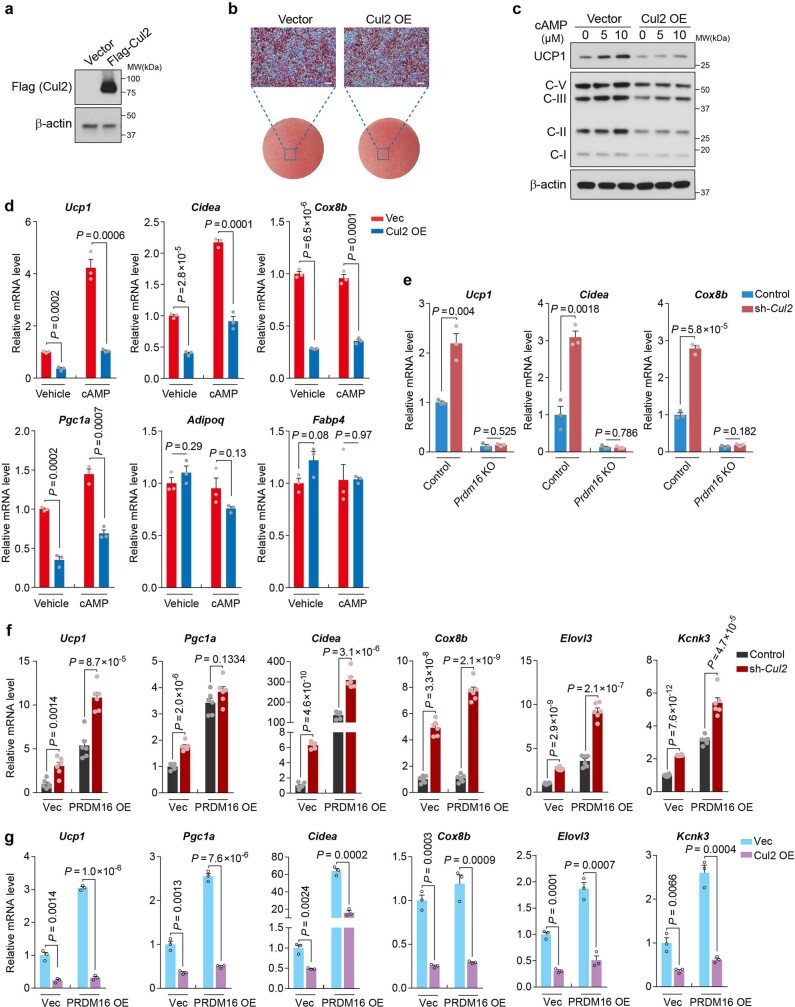
Extended Data Fig. 3. Cul2 overexpression inhibits fat thermogenesis without affecting adipogenesis.
a, Immunoblotting of Cul2 (Flag) proteins in inguinal adipocytes expressing an empty vector or Flag-tagged Cul2 (Cul2 OE). β-actin was used as a loading control. b, Oil-Red-O staining of inguinal adipocytes stably expressing an empty vector or Flag-tagged Cul2 (Cul2 OE). Scale bar, 50 μm. c, Immunoblotting for UCP1 and indicated mitochondrial proteins in inguinal adipocytes stably expressing an empty vector or Flag-tagged Cul2 (Cul2 OE). Differentiated inguinal adipocytes were treated with or without 5 μM or 10 μM forskolin (cAMP) for 8 h. β-actin was used as a loading control. d, Relative mRNA levels of indicated genes in inguinal adipocytes stably expressing an empty vector or Flag-tagged Cul2 in the presence or absence of forskolin (cAMP). n = 3 per group. e, Relative mRNA levels of indicated genes in control and Prdm16 KO inguinal adipocytes expressing a scrambled control shRNA or shRNA targeting Cul2. n = 3 per group. f, Relative mRNA levels of indicated genes in differentiated inguinal adipocytes that expressed Prdm16 (PRDM16 OE) or a control vector and a scrambled control shRNA or shRNA targeting Cul2. n = 6 per group. g, Relative mRNA levels of indicated genes in differentiated inguinal WAT-derived adipocytes expressing Prdm16 (PRDM16 OE) or a control vector and/or Flag-tagged Cul2 (Cul2 OE). n = 3 per group. Representative results in a–c from two independent experiments. Gel source data are presented in Supplementary Fig. 1. d–g, biologically independent samples. Data are mean ± s.e.m.; two-sided P values by unpaired Student’s t-test (d–g).