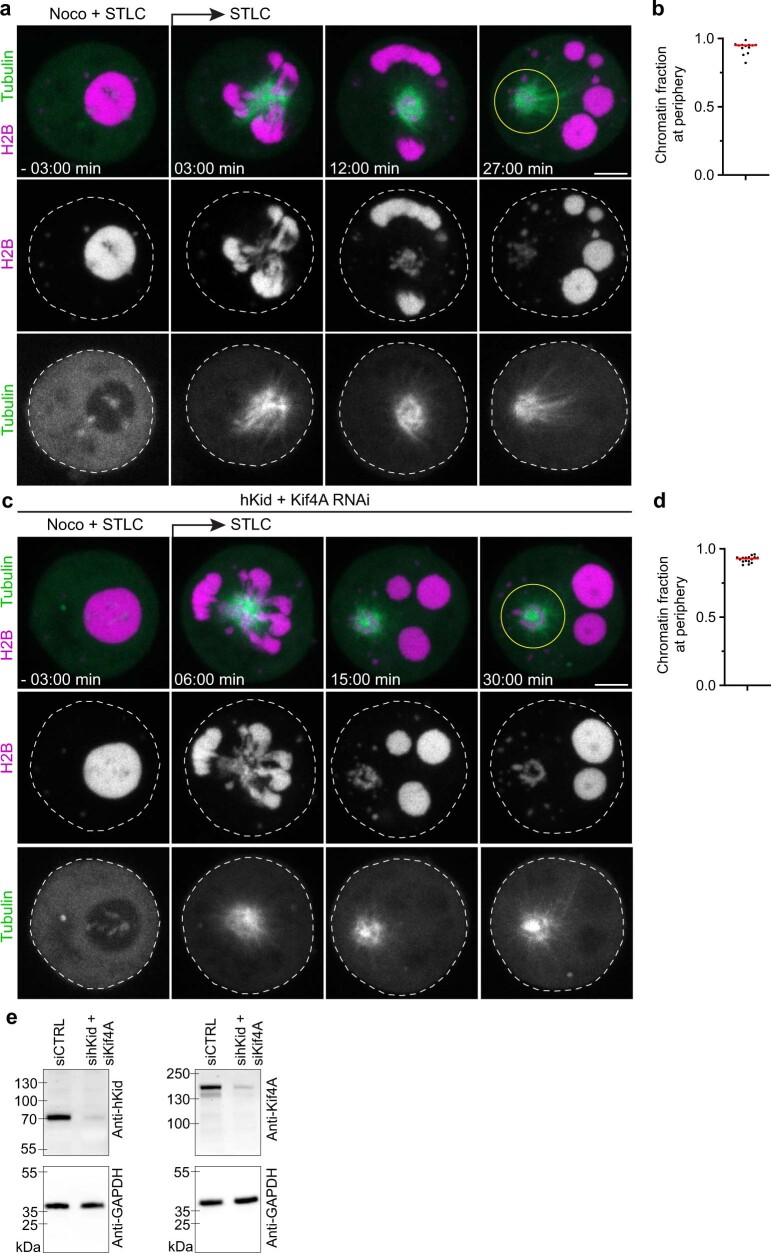
Extended Data Fig. 10. Microtubules push liquified chromatin away from the spindle pole independently of hKid and Kif4A.
a, Time-lapse microscopy of liquified chromatin during chemically-induced assembly of monopolar spindles. Live mitotic HeLa cells expressing H2B-mCherry and eGFP-a-tubulin were treated with nocodazole and STLC and then injected with AluI. Nocodazole was removed at t = 0 min during time-lapse imaging. Projection of 4 z-sections. Representative example of n = 13 cells. b, Quantification of chromatin localization at the cell periphery relative to the region around the spindle monopole at = 36 min for cells as shown in a. c, Time-lapse microscopy of liquified chromatin during spindle assembly as in a for cells depleted of Kid and Kif4a by RNAi. d, Quantification as in b for hKid/Kif4a-RNAi cells. n = 16 cells. Bar indicates mean; significance tested by a two-tailed Mann-Whitney test (P = 0.215) e, Validation of RNAi efficiency by Western Blotting. Samples were collected 30 h after transfection of siRNAs targeting hKid and Kif4a and probed by antibodies as indicated; n = 2 experiments. For gel source data, see Supplementary Figure 1b. Biological replicates: n = 4 (a,b); n = 3 (c,d); n = 2 (e). Scale bars, 5 µm.