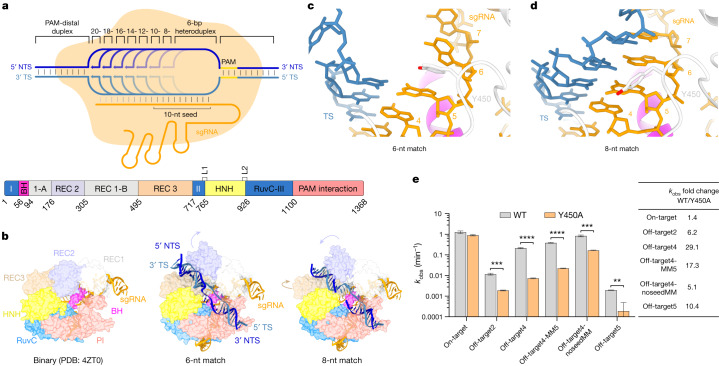
Fig. 1. Target DNA binding induces Cas9 REC lobe restructuring.
a, Top, schematic depicting DNA-bound complexes with increasing extent of complementarity to guide RNA. Bottom, domain composition of SpCas9. 1-A, REC1-A domain; I–III, RuvC domain motifs I–III; BH, bridge helix. b, Structural comparison of the SpCas9 binary (left), 6-nt match (middle) and 8-nt match (right) complexes. c, Zoomed-in view of the seed region of the guide RNA–target DNA heteroduplex in the 6-nt match complex. Tyr450 stacks between the fifth and sixth nucleotide, counting from the PAM-proximal end of the heteroduplex. d, Zoomed-in view of the seed region of the guide RNA–target DNA heteroduplex in the 8-nt match complex. e, Fitted cleavage rate (kobs) of wild-type (WT) and Y450A mutant Cas9 against on-target and off-target substrates. Data represent mean fit ± s.e.m. of n = 4 independent replicates. Two-tailed t-test, ****P < 0.0001, ***P = 0.0002, **P = 0.0011. The P-value for the on-target dataset was not significant (P = 0.1058).