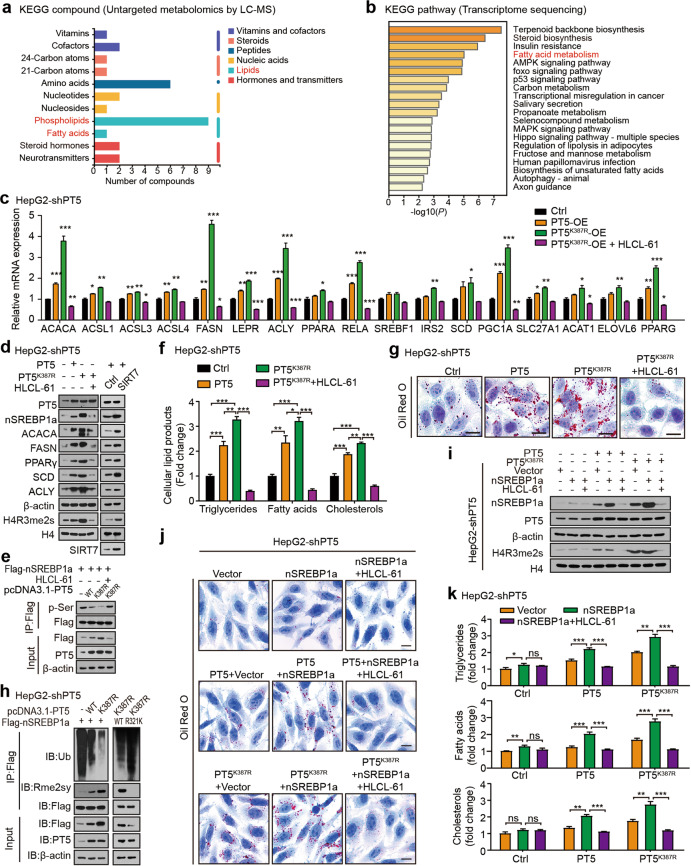
Fig. 4. Desuccinylation of PRMT5 K387 confers lipid metabolism reprogramming by inducing arginine methylation of SREBP1a.
a The KEGG pathway database was used to analyse the metabolites affected by PRMT5 in HepG2 cells. b The KEGG pathway enrichment analysis of PRMT5-regulated target genes in HepG2 cells. c The effect of PRMT5 or PRMT5K387R on the expression of lipid metabolism-associated genes was measured by RT-qPCR analysis in HepG2-shPT5 cells. d The effect of PRMT5, PRMT5K387R and SIRT7 on the protein levels of the indicated proteins was determined by Western blot analysis in HepG2-shPT5 cells. e The effect of PRMT5 and PRMT5K387R on the phosphorylation levels of SREBP1a proteins was tested by IP analysis in HepG2-shPT5 cells. f The levels of intracellular triglycerides, fatty acids and cholesterols were evaluated after the indicated treatments, such as pcDNA3.1-PT5, pcDNA3.1-PT5K387R, or pcDNA3.1-PT5K387R + HLCL-61, in HepG2-shPT5 cells. g The effect of PRMT5 or PRMT5K387R on lipogenesis was determined by Oil Red O staining in HepG2-shPT5 cells. Scale bars, 10 μm. h The ubiquitination and methylation levels of wild-type (WT) SREBP1a and mutant (R321K) SREBP1a were detected by CoIP analysis in HepG2-shPT5 cells transfected with vector, PRMT5 and PRMT5K387R plasmids. i The protein levels of SREBP1a, PRMT5 and H4R3me2s were examined by Western blot analysis in HepG2-shPT5 cells with the indicated treatment. j The effect of PRMT5 or PRMT5K387R with SREBP1a on lipogenesis was determined by Oil Red O staining in HepG2-shPT5 cells. Scale bars, 10 μm. k The levels of intracellular triglycerides, fatty acids and cholesterols were evaluated after the indicated treatment in HepG2-shPT5 cells. Statistically significant differences are indicated: ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA.