Abstract
The aim of our study is to determine the discriminatory power of the Fourier transform infrared spectroscopy (FTIR), using multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) as molecular typing references. The study included seventeen isolates (OXA-23- and OXA-58-producing Acinetobacter baumannii) previously recovered from clinical specimens during the period May 2010–April 2011. Molecular typing was performed by PFGE and MLST. The specimens were analyzed in quadruplicate using the IR Biotyper (Bruker GmbH, Bremen, Germany). For each isolate, the average of the spectra was used for the analysis of the data. Comparing FTIR data with MLST, the results obtained by IR Biotyper are very consistent with those from MLST, since the software was able to differentiate the three ST assigned to the strains. Comparing FTIR data with PFGE, most results could be confirmed, as IR Biotyper clearly differentiated ST-80 SLV OXA-58-producing A. baumannii (pulsotype 3) from the rest of strains of OXA-58-producing A. baumannii (pulsotypes 1 and 2). All the OXA-23-producing A. baumannii isolates (pulsotype 4) grouped together by FTIR. FTIR proved to be an effective tool to investigate local epidemiology, and can achieve the same typeability and discriminatory power as genome-based methods.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-022-00774-6.
Keywords: Acinetobacter baumannii, Fourier transform infrared spectroscopy, MLST, PFGE, Surveillance, Nosocomial
Introduction
Acinetobacter baumannii behaves like an opportunistic pathogen with few virulence factors, which usually produces colonization more often than infection. However, the appearance of fulminant community pneumonias suggests that this microorganism can be highly pathogenic.
The most important role of these bacteria is as cause of nosocomial pneumonia [1]. A. baumannii is described mainly in the form of outbreak, and usually acquires mechanisms of resistance to antibiotics [2]. The most important feature of A. baumannii at the hospital level is the appearance of multidrug-resistant endemic and epidemic strains, usually introduced into the hospital through a colonized patient [3]. The appearance of multi-resistant and even pan-resistant strains occurs specially in intensive care units. It is also possible that they circulate in the community in small proportions and that they are selected in hospitals as a result of the selective pressure of antimicrobial use [4].
Infection control aims to limit the spread of multidrug-resistant bacterial strains. Such outbreaks could be controlled with enhanced hygiene control measures that are quickly implemented after early detection of cross-transmissions [2].
Recently, a new automated typing system based on Fourier transform infrared spectroscopy was commercialized, IR Biotyper (Bruker GmbH, Bremen, Germany). Fourier transform infrared spectroscopy (FTIR) generates spectra based on the absorption of infrared light by the different chemical components of the bacterial cell. Therefore, each microorganism has a highly specific infrared absorption signature correlated with genetic information that allows the identification of microorganisms at the subspecies level [5].
The aim of our study is to determine the discriminatory power of the FTIR, using multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) as molecular typing references, by analyzing an outbreak caused by OXA-23-producing Acinetobacter baumannii that occurred between 2010 and 2011 in Hospital Universitario Puerta del Mar, Cádiz, Spain.
Materials and methods
Bacterial strains
The study included seventeen isolates previously recovered from clinical specimens during a nosocomial outbreak that occurred in the period May 2010–April 2011, and stored at − 80 °C. All isolates were preliminary identified as A. baumannii by using the Wider I system (Soria Melguizo Madrid, Spain) and by matrix-assisted laser desorption/ionization time of flight mass spectrometry MALDI-TOF (Bruker Daltonics, GmbH, Bremen Germany). Partial DNA sequencing of rpoB confirmed the identification as A. baumannii. Multiplex PCR assays were used to detect the following beta-lactamase genes: blaOXA-51-like, blaOXA-23-like, blaOXA-24/40-like, blaOXA-58-like, blaKPC-like, blaGES-like, blaIMP-like, blaVIM-like, and blaNDM-like [6].
Description of the outbreak: molecular genotyping
Before the introduction of an OXA-23-producing A. baumannii in May 2010, there was an endemic setting of nosocomial transmission of OXA-58-producing A. baumannii in the intensive care unit of the Hospital Universitario Puerta del Mar. During the period from May 2010 to April 2011, both clones (OXA-23- and OXA-58-producing) of A. baumannii coexisted. Molecular typing was performed by PFGE, and pulsotypes were determined according to international recommendations [6]. MLST was performed on the different pulsotypes according to Bartual [7].
All OXA-58-producing A. baumannii isolates (n = 8) were classified as ST-2 except one that showed a single-locus variant of ST-80 (ST-80 SLV) (data not published). OXA-23-producing A. baumannii (n = 10) were assigned to ST-1 [6].
A single pulsotype was obtained for OXA-23-producing A. baumannii and three pulsotypes for OXA-58-producing A. baumannii (data not published).
Characteristics of the strains included in the study are presented in Table 1.
Table 1.
Characteristics of the strains included in this study
| Isolate | Source | Isolation date | MLST | PLS | Resistance mechanism |
|---|---|---|---|---|---|
| Abau57 | Blood | 17/02/10 | ST-2 | 2 | OXA-58 |
| Abau86 | Blood | 23/03/10 | ST-2 | 2 | OXA-58 |
| Abau29 | Tracheal aspirate | 26/04/10 | ST-1 | 4 | OXA-23 |
| Abau98 | Central catheter | 26/04/10 | ST-2 | 2 | OXA-58 |
| Abau6 | Tracheal aspirate | 04/05/10 | ST-1 | 4 | OXA-23 |
| Abau180 | Blood | 08/08/10 | ST-1 | 4 | OXA-23 |
| Abau132 | Central catheter | 28/09/10 | ST-1 | 4 | OXA-23 |
| Abau133 | Tracheal aspirate | 28/09/10 | ST-1 | 4 | OXA-23 |
| Abau135 | Tracheal aspirate | 04/10/10 | ST-1 | 4 | OXA-23 |
| Abau140 | Tracheal aspirate | 11/10/10 | ST-1 | 4 | OXA-23 |
| Abau151 | Blood | 21/10/10 | ST-2 | 1 | OXA-58 |
| Abau153 | Bronchoalveolar lavage | 26/10/10 | ST-2 | 1 | OXA-58 |
| Abau146 | Tracheal aspirate | 11/10/10 | ST-1 | 4 | OXA-23 |
| Abau168 | Urine | 29/10/10 | ST-80 SLV | 3 | OXA-58 |
| Abau181 | Urine | 20/12/10 | ST-1 | 4 | OXA-23 |
| Abau187 | Tracheal aspirate | 12/01/11 | ST-2 | 1 | OXA-58 |
| Abau211 | Tracheal aspirate | 28/03/11 | ST-1 | 4 | OXA-23 |
| Abau217 | Sputum | 19/04/11 | ST-2 | 1 | OXA-58 |
PLS, pulsotype
Sample preparation for FTIR spectroscopy
Isolates were grown at 37 °C on Mueller–Hinton II agar (Becton Dickinson, Heidelberg, Germany) for 24 h. Samples were prepared according to the IR Biotyper manufacturer’s instructions. An inoculation loopful (approximately 1 µl) of bacteria was suspended in 50 µl of 70% ethanol solution. Homogenization was obtained by vortexing the sample with metal beads (Bruker, Bremen, Germany) in 1.5-ml microcentrifuge tubes. An additional volume (50 µl) of deionized water was added to obtain a solution with a final volume of 100 µl. An aliquot (15 µl) of the suspension was placed on a sample plate device (Bruker, Bremen, Germany) and dried at 35 °C for 30 min. The pellets were visualized to check for the absence of air bubbles before analysis.
The quality control Infrared Test Standards (IRTS 1 and IRTS 2) of the IR Biotyper Kit were resuspended in 90 µl of deionized water and 90 µl of absolute ethanol which were added and mixed. Twelve microliters of suspension was spotted in duplicate onto the IR Biotyper target, and let dry as described for the samples.
Cluster analysis
Principal component analysis (PCA) was performed applying the IR Biotyper Software version 3.0. The number of principal components used was 3 (95% variance). The specimens were analyzed in quadruplicate using the IR Biotyper with the default analysis settings, and only the region that corresponds to the region of carbohydrates was used. Spectra acquisition, visualization, and processing were performed in transmission mode in the spectral range 4000–600 cm−1 (mid-IR). The second derivative of the spectra was calculated using the Savitzky–Golay algorithm over nine data points. Spectra were then cut to 1300–800 cm−1 and vector-normalized to amplify differences between isolates, and correct variations related to spectra acquisition. The measurement for background and sample measurement was transmission; then, an absorption spectrum was calculated and in IR Biotyper version 3.0 was post-processed by the “make compatible scalar” function of OPUS 8.2.28 software (Bruker GmbH, Bremen, Germany), which interpolates exactly one data point per wavenumber. The resolution of the measurement, when using the default procedures, was 6 cm−1. For each isolate, the average of the spectra was used for the analysis of the data.
Results
Cluster analysis by FTIR
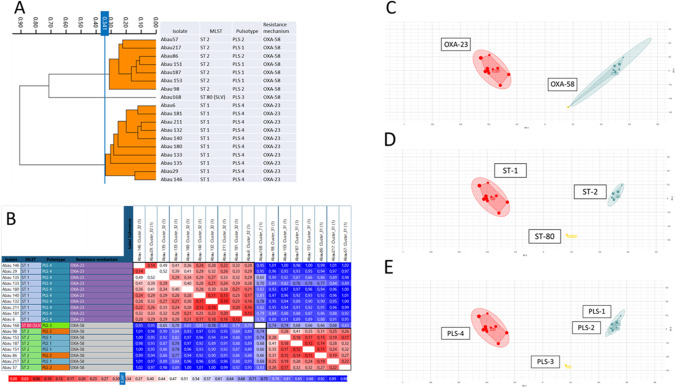
FTIR showed that ST-2 OXA-58-producing A. baumannii were grouped in a large cluster, and OXA-23-producing A. baumannii in a second large cluster, with a cutoff of 0.341. One isolate appeared different in FTIR, ST-80 SLV OXA-58-producing A. baumannii (cutoff of 0.75) (Fig. 1).
Fig. 1.
a Dendrogram of all A. baumannii isolates (cutoff 0.341). b, c, d PCA plot showing the clustering. Analysis by PCA in two dimensions successfully grouped according to the mechanism of resistance (b), ST (c), and pulsotype (d). Number of principal components used: 3 (95% variance). e Distance matrix by Euclidean average linkage (cutoff 0.341)
Comparing FTIR data with MLST, the results obtained by IR Biotyper are very consistent with those from MLST, since the software was able to differentiate the three ST assigned to the strains (Fig. 1).
Comparing FTIR data with PFGE, most results could be confirmed. IR Biotyper clearly differentiated ST-80 SLV OXA-58-producing A. baumannii (pulsotype 3) from the rest of strains of OXA-58-producing A. baumannii (pulsotypes 1 and 2). However, in this last group, FTIR found a high similarity (cutoff = 0.22) between isolates Abau 57 (pulsotype 2) and Abau 217 (pulsotype 1) and also (cutoff = 0.11) between isolates Abau 86 (pulsotype 2) and Abau 151 (pulsotype 1). All the OXA-23-producing A. baumannii isolates (pulsotype 4) grouped together by FTIR.
Discussion
In clinical practice, it is important which method to use for the interpretation of the results in the context of the occurrence of an outbreak, and thus be able to address it as soon as possible. The method used must be reproducible, with high discrimination, fast, and not too expensive.
IR spectroscopy is particularly attractive for the identification and classification of microbes as it is capable of monitoring strain typing. First studies using FTIR spectroscopy focused on foodborne pathogens [8]. Later, the use expanded to other pathogens with clinical or epidemiological importance. FTIR spectroscopy is a fast and cost-effective method to predict the capsular serotype of pneumococci, and probably for typing of other encapsulated species [9]. There are a few studies describing the use of Bruker IR Biotyper for outbreak investigation [9, 10] but, at our knowledge, this is the first one that focused on an A. baumannii outbreak, comparing FTIR and PFGE results.
In our study, the IR Biotyper proved to be an effective tool to investigate local epidemiology, assessing the clonal relationships between different A. baumannii isolates and providing results consistent with the reference DNA-based methods. It successfully identified an OXA-23-producing A. baumannii outbreak cluster, according to the results by PFGE. The clusters by spectroscopy perfectly matched with sequencing results (MLST).
These results are consistent with those reported by other authors for other multidrug-resistant microorganisms. Martak et al. [2] found that FTIR spectroscopy accurately clustered Pseudomonas aeruginosa, Klebsiella pneumoniae, and Enterobacter cloacae isolates belonging to the same ST, and that the IR Biotyper can quickly detect the spread of clones of these microorganisms. However, for A. baumannii, they found that the optimal cutoff ranged from 0.495 to 0.530 with a lower global concordance with MLST, for which three isolates were misclassified, including one ST2 isolate that did not cluster with the other ST2 isolates. In our study, seven ST2 isolates were included, and all of them clustered together. To establish a reliable cutoff in A. baumannii, further studies with FTIR are needed. Our cutoff value and those reported by Martak et al. are higher than those found in Enterobacteriaceae [2, 10, 11].
Rakovitsky et al. [10] recommend that any laboratory planning to use FTIR biotyping for a given organism should initially validate the technical and analytical setup and results using a molecular or genomic approach. They propose complete concordance with whole genome sequencing (WGS) using the cutoff automatically generated by increasing the number of technical replicates to 12. As a limitation in our study, specimens were analyzed in quadruplicate, following manufacturer instructions but, since WGS was not performed, we cannot confirm complete concordance with this technique.
The main limitation of our study is that it was a retrospective analysis of isolates that were already known to be part of an outbreak. Lombardo et al. [12] applied the FTIRS-based IR Biotyper system for a real-time investigation of a carbapenem-resistant A. baumannii outbreak. IR Biotyper enabled a correct and accurate clustering of all the isolates in comparison with PFGE. The measurement of the samples on different days proved to have no influence in the IR Biotyper analysis. So, FTIR might be a screening tool for early assessment of a possible outbreak and could be integrated into routine surveillance.
We conclude that the FTIR Biotyper can achieve the same typeability and discriminatory power as genome-based methods. The results obtained by infrared spectroscopy (FTIR Biotyper) may offer a different way to identify a nosocomial outbreak very quickly, compared to other molecular techniques normally used. Fast, easy-to-apply, and economical, IR Biotyper can be a powerful tool for subtyping of multidrug-resistant microorganisms in local antimicrobial stewardship program.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Dr. Felipe Fernández-Cuenca, Hospital Universitario Virgen Macarena, is acknowledged for his collaboration in MLST and PFGE typing.
Author contribution
I.G.L., F.G.S., and M.R.I. planned the study. I.G.L. performed the FTIR analysis. I.G.L., F.G.S., and M.R.I. analyzed and evaluated the results. I.G.L. and F.G.S. wrote the manuscript. All authors revised the final version of the manuscript.
Data availability
Raw data were generated at Hospital Universitario Puerta del Mar. Derived data supporting the findings of this study are available from the corresponding author on request.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Publication has been approved by all co-authors as well as by the responsible authorities at the institute where the work has been carried out.
Competing interests
The authors declare no competing interests.
Footnotes
Responsible Editor: Jorge Luiz Mello Sampaio
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Joly-Guillou M-L. Clinical impact and pathogenicity of Acinetobacter. Clin Microbiol Infect. 2005;11:868–873. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 2.Martak D, Valot B, Sauget M, Cholley P, Thouverez M, Bertrand X, et al. Fourier-transform infrared spectroscopy can quickly type gram-negative bacilli responsible for hospital outbreaks. Front Microbiol. 2019;10:1440. doi: 10.3389/fmicb.2019.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kempf M, Rolain J-M. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39:105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, et al. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintelas C, Ferreira EC, Lopes JA, Sousa C (2018) An overview of the evolution of infrared spectroscopy applied to bacterial typing. Biotechnol J 13(1). 10.1002/biot.201700449 [DOI] [PubMed]
- 6.Guerrero-Lozano I, Fernández-Cuenca F, Galán-Sánchez F, Egea P, Rodríguez-Iglesias M, Pascual A. Description of the OXA-23 β-lactamase gene located within Tn2007 in a clinical isolate of Acinetobacter baumannii from Spain. Microb Drug Resist. 2015;21(15–2):217. doi: 10.1089/mdr.2014.0155]. [DOI] [PubMed] [Google Scholar]
- 7.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehling-Schulz M, Svensson B, Guinebretiere MH, Lindback T, Andersson M, Schulz A, et al. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology. 2005;151:183–197. doi: 10.1099/mic.0.27607-0]. [DOI] [PubMed] [Google Scholar]
- 9.Burckhardt I, Sebastian K, Mauder N, Kostrzewa M, Burckhardt F, Zimmermann S. Analysis of Streptococcus pneumoniae using Fourier-transformed infrared spectroscopy allows prediction of capsular serotype. Eur J Clin Microbiol Infect Dis. 2019;38:1883–1890. doi: 10.1007/s10096-019-03622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakovitsky N, Frenk S, Kon H, Schwartz D, Temkin E, Solter E, et al. Fourier transform infrared spectroscopy is a new option for outbreak investigation: a retrospective analysis of an extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. J Clin Microbiol. 2020;58:e00098–e120. doi: 10.1128/JCM.00098-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Zhou H, Lu J, Sun Q, Liu C, Zeng Y, et al. Evaluation of the IR Biotyper for Klebsiella pneumoniae typing and its potentials in hospital hygiene management. Microb Biotechnol. 2020;14:1343–1352. doi: 10.1111/1751-7915.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombardo D, Cordovana M, Deidda F, Pane M, Ambretti S. Application of Fourier transform infrared spectroscopy for real-time typing of an Acinetobacter baumannii outbreak in an intensive care unit. Future Microbiol. 2021;16:1239–1250. doi: 10.2217/fmb-2020-0276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data were generated at Hospital Universitario Puerta del Mar. Derived data supporting the findings of this study are available from the corresponding author on request.
Not applicable.