Fig. 2.
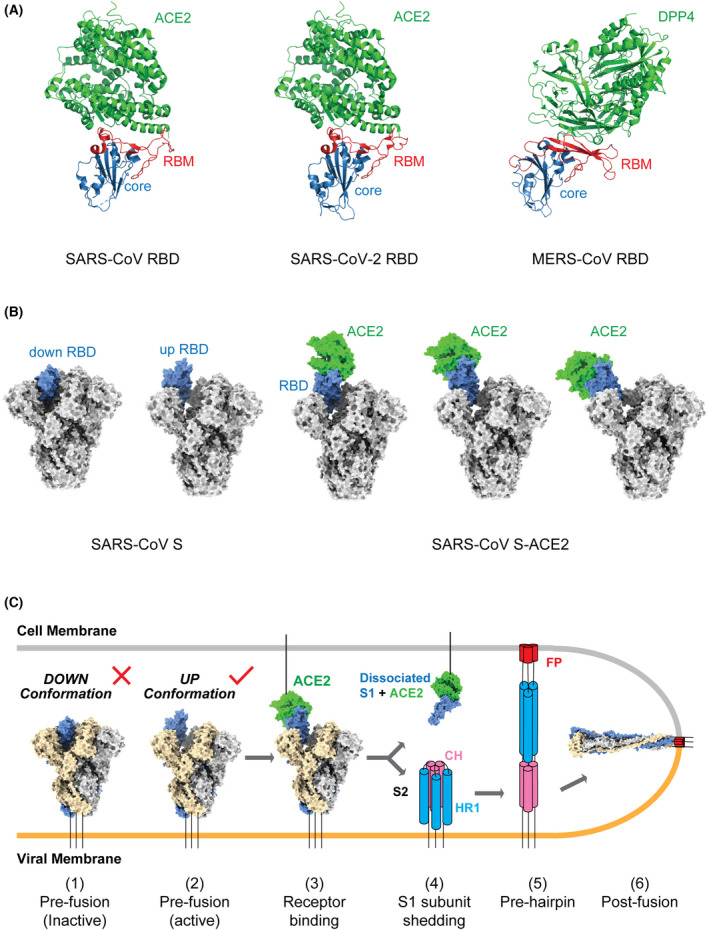
Interaction between CoV S glycoprotein and host receptor. (A) Overall structures of the SARS‐CoV (left) and SARS‐CoV‐2 RBDs (middle) bound to ACE2 and MERS‐CoV RBD (right) bound to DPP4. RBD core domains are coloured blue and RBMs are coloured red. Both the ACE2 and DPP4 receptors are coloured green. PDB codes: SARS‐CoV RBD–ACE2 complex, 2AJF; SARS‐CoV RBD‐2–ACE2 complex, 6M0J; MERS‐CoV RBD–DPP4 complex, 4L72. (B) Overall structures of SARS‐CoV S glycoprotein (left) and ACE2‐bound SARS‐CoV S glycoprotein (right). S glycoprotein is shown in grey and the dynamic RBD in blue. ACE2 is coloured green. PDB codes: 6ACG, 6ACJ, 6ACK, 6ACD and 6ACC. (C) Model of the pre‐fusion to post‐fusion transition of the SARS‐CoV S trimer. The down‐to‐up conformational transition of RBD (1–2) exposes the receptor‐binding site, followed by ACE2 binding (3), ‘opening’ up of the S trimer, shedding of the S1 subunits (4), and a large‐scale conformational change of the unwrapped S2 subunits for FP insertion into the cell membrane (5, pre‐hairpin state) and the formation of a six‐helix bundle to enable membrane fusion (6). This model is based on current knowledge and could be optimized with future data. [Colour figure can be viewed at wileyonlinelibrary.com]
