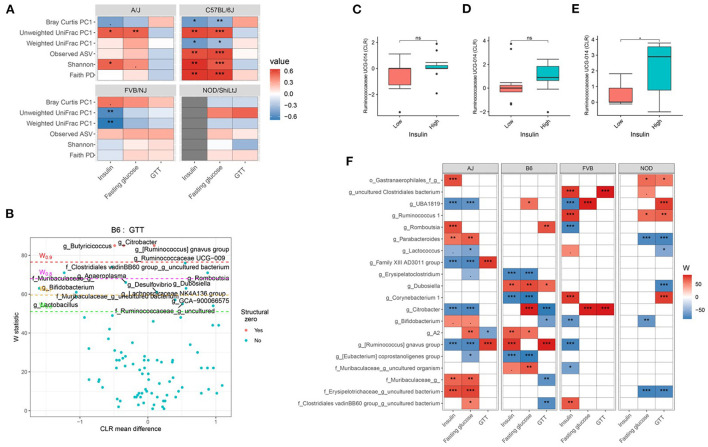
Figure 6.
Host genetics and gut microbiota interact to modulate host glucose metabolism. (A) Association between host glucose metabolism phenotypes and α-diversity or first principal component (PC1) of β-diversity measured by mouse strains determined by using Spearman correlation. As indicated, red represents a positive correlation and blue represents a negative correlation. “***” = adj.p < 0.001, “**” = adj.p < 0.01, “*” = adj.p < 0.05, “.” = adj.p < 0.10. (B) Volcano plot of the differential genera abundance between C57BL/6J mice with high (above median) and low (below median) GTT AUC determined by ANCOM analysis. Structural zero represents the presence of the bacteria in one group and the complete absence in another group. Relative abundance of the Ruminococcaceae UCG-014 in AJ (C) C57BL/6J (D) and FVB/NJ (E) mice with high and low plasma insulin levels. Mean abundance was compared using the Wilcoxon rank-sum test. For (C–E), “*” = P < 0.05 and “ns” = not significant. (F) Differential genera abundance between high (above median) and low (below median) blood glucose-related phenotypes determined by ANCOM analysis. 20 most differentially abundant (based on cumulative ANCOM W-value) were selected for the graph. For easier presentation, ANCOM W values were converted to negative if CLR effect size is negative. Red indicates higher genera abundance in the above-median group and blue bacteria represents higher genera abundance in the below-median group. White represents a non-significant result obtained from ANCOM analysis. Red and blue represent significant associations determined by ANCOM. Models were adjusted for diet and sex as confounding factors. The median was calculated for each mouse strain. ANCOM models were FDR (BH-method) corrected for multiple comparisons and a significant sub-hypothesis test at a level of adj.P < 0.05 were counted toward W value. For (E), “***” = ≥W0.9, “**” = ≥W0.8, “*” = ≥W0.7, and “.” = ≥W0.6.