Abstract
The compound (E)-ethyl 3-(2-(2,4-dinitrophenyl)hydrazono)butanoate (3) was synthesised and crystallized using ethanol as a solvent. The compound was characterized by 1H NMR, and single crystal X-ray diffraction. The compound crystallizes in the monoclinic crystal system with the space group P21/c. The intermolecular interactions and the interaction energies responsible for the stabilization of the molecules were determined by Hirshfeld surface analysis and energy framework calculations. The structure of the compound was optimized by Density Functional Theory calculations and HOMO–LUMO energy gap was calculated. The non–covalent interactions were revealed by reduced density gradient analysis. The Mulliken atomic charges and natural atomic charges were calculated by density functional theory calculations. The reactive sites present in the molecule are shown by molecular electrostatic potential map. The inter and intra molecular charge transfer were investigated by NBO analysis.
Keywords: Schiff base, HOMO-LUMO, Non-covalent interactions, NBO
Highlights
-
•
Crystal structure was confirmed by X-ray diffraction analysis and the atomic coordinates were optimized by DFT calculations.
-
•
Hirshfeld surface analysis and energy frameworks calculations were carried out.
-
•
HOMO - LUMO frontier molecular orbitals and molecular electrostatic potential were studied.
-
•
Reduced density gradient (RDG), and topology analyses were performed.
-
•
Natural bond orbital analysis was performed to study intramolecular charge transfer.
Schiff base; HOMO-LUMO; Non-covalent interactions; NBO.
1. Introduction
With uncountable agricultural and industrial operations, the release of pollutants in the form of cation and anions has escalated the threats to the environment, leading to widespread animal and human health issues [1]. The demand for identification of the pollutants (e.g., toxins and metal ions) is considerable, especially in health and environmental sectors [2, 3]. The Schiff base shows incredible results for metal ion detection in this regard [4]. Schiff bases are molecules that feature an imine or azomethine (–CH = N–) functional group. They serve as ligands in diverse metal complexes. Schiff bases are a common family of organic molecules employed in a variety of domains that include analytical, biological, and inorganic chemistry [5, 6]. Schiff bases can find, and identify aldehydes and ketones. among other things. Preparative use, purification of carbonyl or amino compounds, and shielding of these groups during complex or sensitive processes are a small selection of the applications for Schiff bases [7]. Schiff bases can be employed as pigments/dyes [8], catalysts [9], intermediate compounds, corrosion inhibitors [10], and polymer stabilisers [11] in addition to biological functions [12]. Schiff bases have been successfully employed as highly active and promising sensing materials for a variety of ion sensors including optical, electrochemical, and membrane sensors [13, 14, 15]. Several Schiff bases are shown to be active antifungal agents [16]. They also exhibit anticancer activity [17]. As a result, the researchers are trying to produce new heterocyclic/aryl Schiff bases for the development of environment friendly technologies [18]. Various approaches, such as 1H NMR and elemental analyses, are used to determine and describe the structure of the compound. Single crystal X-ray diffraction method is employed to confirm the 3D structure of the molecule. The computational analysis using DFT calculations are performed to optimize the structure, to calculate HOMO–LUMO energy gap and other global chemical descriptors [19]. Hirshfeld surface analysis unveils stabilizing intermolecular interactions. The inter and intramolecular charge transfer in the molecule is studied by natural bond analysis.
2. Experimental procedure
2.1. Materials and methods
Chemicals were procured from Sigma Aldrich and TCI Chemicals. The melting point was determined using a thermometer and a Chemi Line CL725 Micro Controller-based melting point device. Agilent-NMR spectrophotometer (VNMRS-400 MHz) was employed to record the NMR spectrum in dimethyl sulfoxide (DMSO), while a SynaptG2 spectrophotometer (VG70-70H) was utilized to obtain the mass spectrum.
2.2. Synthesis of (E)-ethyl 3-(2-(2,4-dinitrophenyl)hydrazono)butanoate
The compound (2,4-dinitrophenyl)hydrazine (1, 0.001 mol) was refluxed along with ethyl 3-oxobutanoate (2, 0.001 mol) using ethanol as solvent in the presence of acetic acid for 4 h. The reaction was monitored using TLC with hexane and ethyl acetate (4:1) as solvent system. After the completion of the reaction the reaction mixture was poured on crushed ice. The synthetic procedure is shown in Figure 1.
Figure 1.
Reaction scheme for the preparation of the compound 3.
(E)-ethyl 3-(2-(2,4-dinitrophenyl)hydrazono)butanoate (3): Yield: 85%. Mp. 111-113 °C. 1H NMR (DMSO): δ 1.20 (t, 3H, CH3), 2.10 (s, 3H, =C–CH3), 3.52 (s, 2H, CH2), 4.13 (q, 2H, CH2), 6.92–7.58 (m, 3H, Ar–H), 10.79 (s, 1H, NH) [Figure 2]. LC-MS m/z 311 (M+1). Anal. Calcd for C12H14N4O6: C, 46.45; H, 4.55. Found: C, 46.41; H, 4.52 %.
Figure 2.
1 H-NMR of the compound 3.
2.3. X-ray data collection and reduction
A block shaped crystal of approximate dimensions 0.20⨯0.25⨯0.30 mm was chosen for X-ray intensity data collection. Bruker D8 VENTURE diffractometer with PHOTON II detector was employed for data collection operated at 296 K using MoKα radiation of wavelength 0.78Å. The data set was processed using SAINT PLUS [20]. The structure was solved by direct methods and refined by full-matrix least squares method on F2 using SHELXS and SHELXL programs [21]. All the hydrogen atoms were placed at chemically acceptable positions and refined. 210 parameters were refined with 4174 distinct reflections which converged R1 to 0.0616 (wR2 = 0.1685) with goodness-of-fit on F2 = 1.017. PLATON [22] was used to perform geometrical calculations. Molecular packing diagrams and ORTEP were generated using MERCURY [23].
2.4. Computational details
The molecules in the crystals are linked up by the intermolecular interactions. The interactions are unveiled by Hirshfeld surface analysis by plotting dnorm, shape index, curvedness and electrostatic potential. CrystalExplorer 17.5 [24] software was utilized to analyse Hirshfeld surface, and to generate 2D fingerprint plots. The interactions in the crystal were envisaged on the Hirshfeld surface using red-white-blue colours [25]. 3D energy frameworks and interaction energies between molecular pairs were calculated by CrystalExplorer 17.5 [26].
Density Functional Theory (DFT) calculations using B3LYP, 6-311G (d, p) basis set in gas phase of the compound 3 were performed using Gaussian 09 [27]. The frontier molecular orbitals (HOMO and LUMO) are visualized using Gaussview 6 [28]. The RDG to analyze non–covalent interactions, topology analysis to study intra molecular interactions, analysis of charge distribution by MEP map were performed using Multiwfn 3.8 [29] and visualized using Visual Molecular Dynamics [30].
3. Results and discussion
3.1. Structure analysis
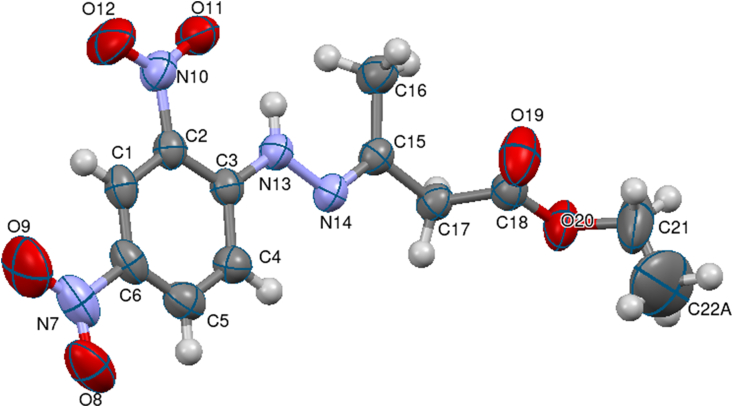
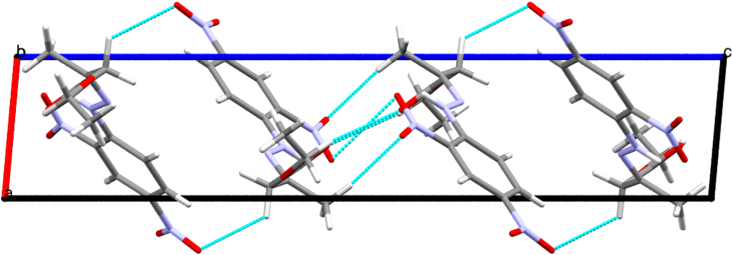
The compound 3 crystallizes in the monoclinic crystal system with the space group P21/c. The cell parameters are a = 4.5147(6) Å, b = 14.817(2) Å, c = 22.461(3) Å, and β = 95.061(5)°. Table S1 summarizes the crystal data and structure refinement details. Figures 3 and 4 portray the ORTEP, and packing along the a-axis of the compound 3 respectively. Tables S2, S3, and S4 gives the bond lengths, bond angles and torsion angles of the compound 3.
Figure 3.
ORTEP of the molecule with thermal ellipsoids drawn at 50% probability.
Figure 4.
Packing of the molecules viewed down the b-axis.
The carbon atoms (C1–C2–C3–C4–C5–C6) of dinitrobenzene ring in the compound are sp2 hybridized. The C1 atom shows maximum deviation from the mean plane with r.m.s value -0.003(2). The bond angle and torsion angle values C2–C3–C4 = 117.1°, C3–C4–C5 = 121.1°, C6–C1–C2–C3 = 0.3°, C3–C4–C5–C6 = 0.1° of the dinitrobenzene ring are signifying nearly planar trigonal geometry. The nitro groups attached to the benzene ring O8–N7–O9 and O11–N10–O12 have bent geometry with the bond angles 122.0°and 124.1°. The torsion angle values O9–N7–C6–C5 (−175.3°) and O11–N10–C2–C1 (−175.4°) indicate the -anti-periplanar conformation of the ring. The ethoxy group is non-planar with the dinitrobenzene ring with a dihedral angle = 85.65°. The molecules are linked by C–H⋯π, N–O⋯π, and intramolecular interactions of the type N–H⋯O, N–H⋯N, C–H⋯N and C–H⋯O. Hydrogen bond interaction and N–O⋯π geometry are listed in Tables 1 and 2.
Table 1.
Hydrogen bond geometry.
| Atoms | D-H (Å) | H⋯A (Å) | D-A (Å) | D-H⋯A (◦) |
|---|---|---|---|---|
| N13–H13⋯O11 | 0.86 | 1.99 | 2.613(3) | 128 |
| N13–H13⋯N10 | 0.86 | 2.63 | 2.923(3) | 102 |
| C4–H4⋯N14 | 0.93 | 2.41 | 2.734(3) | 100 |
| C21–H21B⋯O19 | 0.97 | 2.18 | 2.652(4) | 108 |
Table 2.
N–O ⋯π interaction involved in the molecular structure.
| N⋯O | CgJ | O⋯Cg(Å) | O⊥ (Å) | γ (°) | N–O⋯cg (°) | N⋯cg (Å) | N–O⋯π (°) |
|---|---|---|---|---|---|---|---|
| N7⋯O8 | Cg1i | 3.339(3) | −3.265 | 12.03 | 84.27(17) | 3.439(3) | 5.78 |
| N7⋯O9 | Cg1i | 3.881(3) | −3.476 | 26.42 | 60.04(17) | 3.439(3) | 4.12 |
i = 1+x, y, z.
3.2. Intermolecular interactions and energies
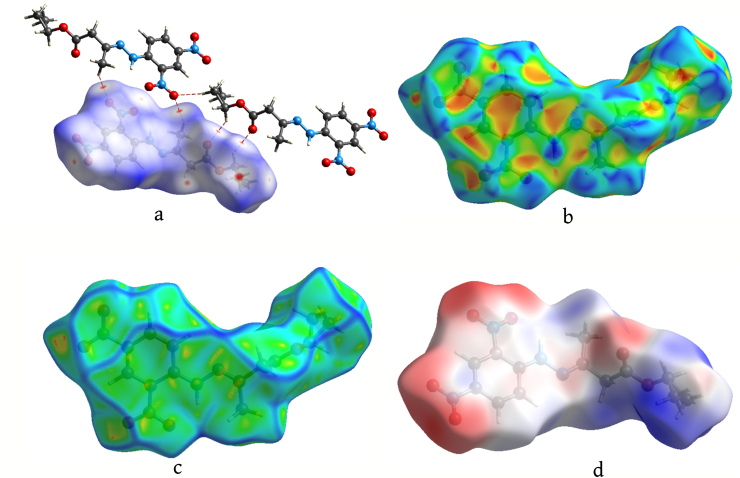
On the Hirshfeld surface (dnorm) the red spots depict the short interactions present in the molecule (Figure 5a). The quantitative measures of the Hirshfeld surface are total volume (367.14 Å3), area (348.54 Å2), globularity (0.711), and asphericity (0.347). The value of globularity is < 1.0 which indicates that the molecular surface is more structured. The structural anisotropy of compound 3 is given by asphericity value [31].
Figure 5.
Hirshfeld surface mapped for (a) d norm (b) Shape index (c) Curvedness (d) Electrostatic potential.
The shape index and curvedness mapped on Hirshfeld surface render information on other weak interactions present in the compound. On the shape index surface (Figure 5b) the C–H⋯π and N–O⋯π interactions are indicated as red regions around the participating atoms. The π-π interactions in the molecule are indicated by flat regions on the curvedness surface (Figure 5c).
The electrophilic and nucleophilic regions are identified by electrostatic potential map plotted on Hirshfeld surface using DFT-B3LYP/6-31G(d, p) hybrid functional. The red coloured regions around oxygen atoms (Figure 5d) show that they are good electron acceptors [32].
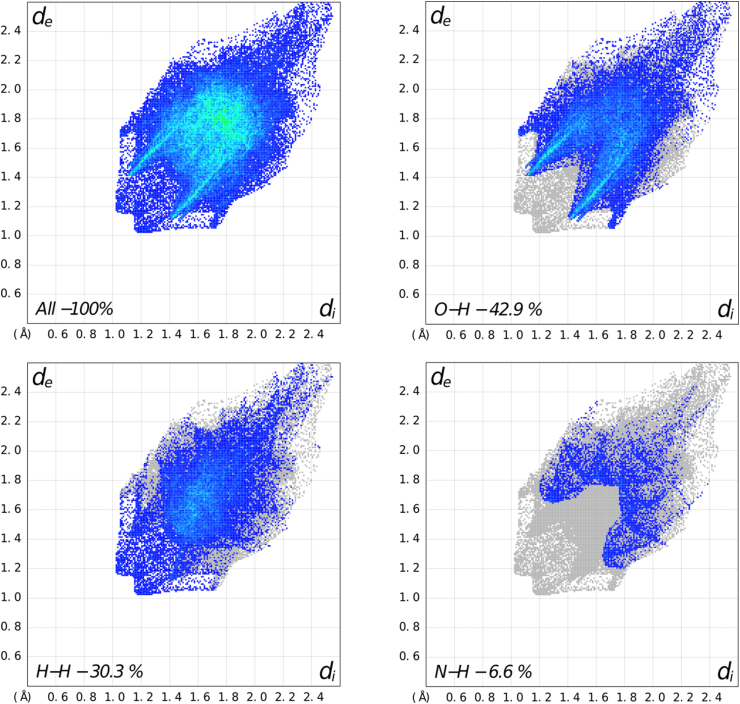
Two dimensional fingerprint plots show specific contribution to the total Hirshfeld surface area. The selected decomposed fingerprint plots are shown in Figure 6. The significant contributions are from the contacts O–H (42.9%) and H–H (30.3%). Other specific interactions are listed in Table 3 [33].
Figure 6.
Fingerprint plots showing percentage contribution to the total Hirshfeld surface of compound 3.
Table 3.
Percentage contribution to the total Hirshfeld surface area.
| Intermolecular Contacts | Contribution (%) |
|---|---|
| O–H | 42.9 |
| H–H | 30.3 |
| N–H | 6.6 |
| H–C | 5.5 |
| C–O | 5.0 |
| N–C | 3.4 |
| O–O | 3.7 |
Pair wise interaction energies for the molecule 3 within a crystal were calculated. The total interaction energy (−209 kJ mol−1) is composed of four components electrostatic (−86.03 kJ mol−1), polarization (−29.15 kJ mol−1), dispersion (−174.02 kJ mol−1), and repulsion energy (79.96 kJ mol−1). The individual components of energy and total energy for the specific molecular pairs are given in Table 4. The molecular pairs involved in the interaction energy calculation are shown in Figure s1. For the molecule under discussion dispersion energy is dominant. The energy frameworks which visualize the strength of interaction energy are shown in Figure 7 for Coulombic energy, dispersion energy, and total energy with red, green, and blue colours respectively. The radii of cylinders connecting the centroid of the molecules represent relative interaction energy strengths [34].
Table 4.
Interaction energies of the molecular pairs involved in energy calculation in kJ/mol. R is the distance between molecular centroids in Å and N is the number of molecular pairs involved.
Figure 7.
Energy frameworks constructed for Coulomb energy, dispersion energy and total energy.
Where E_ele is the electrostatic energy, E_pol is the polarization energy, E_dis is the dispersion energy, E_rep is the repulsive energy and E_tot is the total energy.
3.3. HOMO – LUMO and quantum chemical descriptors
Highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) play a significant role in the determination of chemical behavior of the molecule. The LUMO is regarded as electrophile, and HOMO as nucleophile which is a electron donor. The enegy gap between HOMO and LUMO is significant in determining the chemical stability of the molecule [35]. The HOMO and LUMO values of the molecule 3 and other global chemical descriptors were calculated and are listed in Table 5. A molecule exhibits good chemical reactivity due to the smaller energy gap value. A molecule with high energy gap has good kinetic stability [36]. The energy gap of the molecule 3 is found to be 3.647 eV Figure 8 displays the localization of molecular orbitals in the molecule 3. HOMO and LUMO are localized on the dinitrobenzene ring.
Table 5.
HOMO, LUMO, and quantum chemical descriptor values of the molecule 3.
| Parameter | Value |
|---|---|
| Elumo | −2.990 eV |
| Ehomo | −6.637eV |
| ΔE | 3.647 eV |
| Ionization potential (I) | 6.637 eV |
| Electron affinity (A) | 2.990 eV |
| Chemical potential μ | −4.813 eV |
| Chemical Hardness η | 1.823 eV |
| Electronegativity χ | 4.813 eV |
| Electrophilicity ω | 6.353 eV |
| Global softness σ | 0.548 eV−1 |
Figure 8.
HOMO and LUMO of the molecule 3.
3.4. Atomic charge analysis
3.4.1. Mulliken and natural atomic charge
For the molecule 3 both Mulliken charge and natural atomic charge analyses were carried out via DFT methods [37]. Both the charges on each atom are listed in Table 6. All the hydrogen atoms show positive charge since they are electron donors. C18 atom possesses highest electropositive charge due to the adjacent electronegative atom O19. The atoms O20, O19, C22, C17, C16 possess high negative charge compared to other atoms. The distribution of atomic charges are shown in Figure S2.
Table 6.
Mulliken and Natural atomic charges of the molecule.
| Atoms with their number | Mulliken charge | Natural atomic charge | Atoms with their number | Mulliken charge | Natural atomic charge |
|---|---|---|---|---|---|
| C4 | −0.11 | −0.23 | C18 | 0.37 | 0.85 |
| C3 | 0.3 | 0.22 | O19 | −0.34 | −0.6 |
| C5 | −0.03 | −0.14 | O20 | −0.33 | −0.56 |
| C6 | 0.1 | 0.03 | C21 | −0.03 | −0.02 |
| C1 | −0.02 | −0.15 | C22 | −0.3 | −0.59 |
| C2 | 0.1 | 0.02 | H4 | 0.13 | 0.24 |
| N7 | 0.18 | 0.51 | H5 | 0.14 | 0.24 |
| O8 | −0.27 | −0.39 | H1 | 0.17 | 0.27 |
| O9 | −0.27 | −0.39 | H13 | 0.28 | 0.42 |
| N10 | 0.18 | 0.51 | H16A | 0.14 | 0.22 |
| O11 | −0.33 | −0.37 | H16B | 0.16 | 0.24 |
| O12 | −0.25 | −0.44 | H16C | 0.13 | 0.22 |
| N13 | −0.39 | −0.38 | H17A | 0.16 | 0.24 |
| N14 | −0.17 | −0.39 | H17B | 0.16 | 0.23 |
| C15 | 0.11 | 0.31 | H21A | 0.13 | 0.18 |
| C16 | −0.32 | −0.66 | H21B | 0.13 | 0.18 |
| C17 | −0.26 | −0.51 | H22A | 0.12 | 0.2 |
| H22C | 0.12 | 0.21 |
3.4.2. Molecular electrostatic potential (MEP)
The molecular electrostatic potential of 3 is visualized in Figure 9. The MEP map allows us to understand the reactive sites, and the relative polarity of the molecule. In molecule 3 the potential ranges from −3.3902e + 01 kcal/mol to 2.736e + 01 kcal/mol. The potentials of the molecule are displayed on the map with increasing order using red < yellow < green < blue colours [38]. The red coloured regions encompassing the oxygen atoms corresponding to low potential values infer that those atoms exhibit electrophilic nature. The hydrogen atoms encompassed by extreme blue coloured regions suggest their nucleophilic nature.
Figure 9.
MEP map of the molecule 3.
3.5. Thermodynamic properties
On the basis of vibrational analysis at B3LYP/6-311 G (d, p) the thermodynamic parameters were calculated. The variation of entropy (S), heat capacity (C), and enthalpy (H) with temperature are listed in Table 7 and shown in Figure 10. One can observe that the thermodynamic parameters increase with temperature. Total energy, specific heat at constant volume, entropy due to vibration, rotation, translation and zero point vibrational energy are calculated and are listed in Table 8. The correlation equations between entropy (S), heat capacity (Cp), and enthalpy (H) with temperature obtained by quadratic fitting [39] are
| Cp = −0.00045T2 + 1.0879T + 50.239 (R2 = 0.999) |
| H = 0.0003 T2 + 0.16537 T −13.7959 (R2 = 0.999) |
| S = −0.00036T2 + 1.35166T + 303.66 (R2 = 0.999). |
Table 7.
Thermodynamic functions at different temperature for the molecule 3.
| Temperature (K) | S (J/mol K) | C (J/mol K) | H (kJ/mol) |
|---|---|---|---|
| 100 | 427.23 | 158.8 | 10.26 |
| 200 | 564.39 | 246.08 | 30.55 |
| 298.15 | 678.5 | 331.1 | 58.87 |
| 300 | 680.55 | 332.684 | 59.49 |
| 400 | 787.698 | 414.65 | 96.93 |
| 500 | 888.033 | 485.08 | 142.03 |
| 600 | 981.757 | 542.796 | 193.52 |
| 700 | 1069.08 | 589.715 | 250.22 |
| 800 | 1150.41 | 628.18 | 311.18 |
| 900 | 1226.3 | 660.080 | 375.64 |
| 1000 | 1297.27 | 686.82 | 443.03 |
Figure 10.
Variation of the thermodynamic parameters with temperature.
Table 8.
Theoretically computed thermodynamic parameters of the molecule 3.
| Parameters | Values | |||
|---|---|---|---|---|
| Zero point vibrational energy (kcal mol−1) | 169.250 | |||
| Total | Translational | Rotational | Vibrational | |
| Energy (k cal mol−1) | 182.729 | 0.889 | 0.889 | 180.952 |
| Specific heat at constant volume (cal mol−1 K−1) | 77.146 | 2.981 | 2.981 | 71.185 |
| Entropy (cal mol−1 K−1) | 162.138 | 43.091 | 35.499 | 83.548 |
3.6. Non-covalent interactions (NCI)
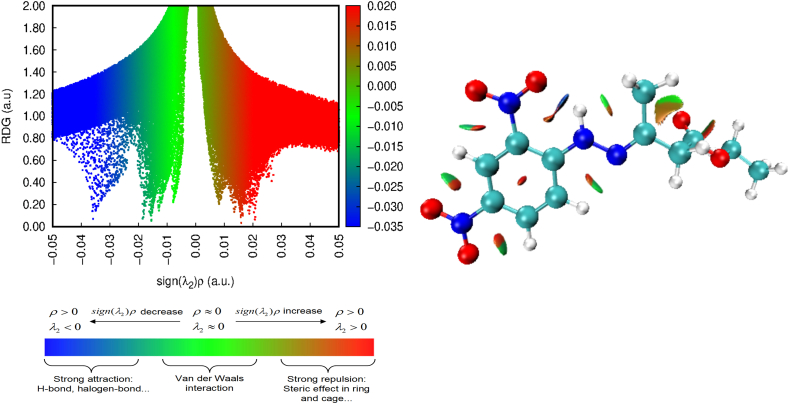
The structure of the molecule is influenced by covalent, noncovalent, and electrostatic interactions. The NCI is revealed by Reduced Density Gradient analysis (RDG). NCI is visualized by gradient isosurfaces coloured according to the strength of the interaction. Sign(λ2)ρ values indicate the type of bonding— where large negative values specify hydrogen bonding, while large positive values specify repulsive interactions. The weak van der Waals interactions are specified by the values of sign(λ2)ρ near zero [40]. The scattered plot and isosurfaces of the molecule 3 are shown in Figure 11. The red colored isosurface is a clear indication of the steric repulsion present in the benzene ring with sign(λ2)ρ = 0.02. The molecule possesses strong hydrogen bond (N–H⋯O) shown as blue isosurface with sign(λ2)ρ value nearly -0.04. All the green isosurfaces depict the weak van der Waals interactions in the molecule 3 [41].
Figure 11.
NCI and scattered plot of the molecule 3.
3.7. Topology analysis
Topological analysis of atoms in the molecule was performed to look into the intra-molecular interactions in the molecule 3. According to Atoms in Molecule (AIM) analysis the presence of bond critical path (BCP) ensures that the bond between the atoms. BCP generated for the molecule 3 is shown in Figure 12. The BCP corresponding to N13–H13⋯O11 validates the presence of intramolecular interaction observed in X-ray diffraction study. The total electron density ρ(r) and Laplacian (∇2ρ) describe the nature of bonds. Total electron density is given by the expression,
Figure 12.
Bond critical path generated (yellow line) for the molecule 3.
where G(r) and V(r) are Lagragian kinetic energy and potential energy densities at critical points, respectively. The BCP with ∇2ρ(rBCP) = 0.1354 and G(rBCP) (0.032) greater than V(rBCP) (−0.0311) indicates the existence of hydrogen bond. The values ∇2ρ(r)BCP > 0 and G(rBCP) +V(rBCP) > 0 indicate that the bond is a weak hydrogen bond. Ratio of −G(r BCP)/V(r BCP) = 1.045 indicates that the hydrogen bond has a non-covalent nature [42].
3.8. Natural bond analysis
Natural bond orbital (NBO) analysis is an efficient method to understand the inter and intramolecular interactions. The delocalization of electron density within the molecule and intramolecular charge transfer are investigated from NBO analysis. Donor-acceptor interactions in the NBO of the molecule 3 were carried out with the second order perturbation theory analysis in the Fock matrix. Interaction among atoms and their charge transfer is investigated. Table 9 lists the second-order perturbation energies of the molecule 3. The strong interactions are through the atoms having higher stabilization energy E(2) [43]. In this molecule, maximum stabilization energy is found in the bond LP (3) O8 - π∗ (N7 – O9).
Table 9.
Selected second-order perturbation energies of the molecule 3.
| Donor | Occupancy | Acceptor | Occupancy | E(2) kcal/mol | E(j)-E(i) a.u. | F(i, j) a.u. |
|---|---|---|---|---|---|---|
| LP (3) O8 | 1.45 | π∗ (N7–O9) | 0.64 | 162.72 | 0.14 | 0.14 |
| π∗ (C2–C6) | 0.49 | π∗ (C4–C5) | 0.34 | 134.4 | 0.03 | 0.08 |
| LP (3) O11 | 1.51 | π∗ (N1–O12) | 0.63 | 133.7 | 0.15 | 0.13 |
| π∗ (C2–C6) | 0.49 | π∗ (C1–C3) | 0.24 | 85.12 | 0.03 | 0.08 |
| LP (1) N13 | 1.62 | π∗ (C2–C6) | 0.49 | 55.7 | 0.26 | 0.11 |
| LP (2) O25 | 1.79 | π∗ (C21–O24) | 0.22 | 48.79 | 0.34 | 0.12 |
| π (C2–C6) | 1.59 | π∗ (N10–O12) | 0.63 | 33.63 | 0.15 | 0.07 |
| LP (2) O24 | 1.85 | σ∗ (C21–O25) | 0.1 | 31.2 | 0.65 | 0.13 |
| π (C2–C6) | 1.59 | π∗ (C4–C5) | 0.34 | 26.52 | 0.3 | 0.08 |
| LP (1) N13 | 1.62 | π∗ (N14–C15) | 0.17 | 24.93 | 0.29 | 0.08 |
| π (C4–C5) | 1.68 | π∗ (N7–O9) | 0.64 | 24.55 | 0.16 | 0.06 |
| π (C1–C3) | 1.72 | π∗ (C2–C6) | 0.49 | 23.54 | 0.26 | 0.07 |
| π (C4–C5) | 1.68 | π∗ (C1–C3) | 0.24 | 21.65 | 0.3 | 0.07 |
| π∗ (N10–O12) | 0.05 | π∗ (C2–C6) | 0.49 | 20.37 | 0.12 | 0.06 |
| LP (2) O24 | 1.85 | σ∗ (C20–C21) | 0.07 | 19.89 | 0.63 | 0.1 |
| LP (2) O12 | 1.89 | σ∗ (N10–O11) | 0.06 | 19.63 | 0.69 | 0.11 |
| LP (2) O9 | 1.9 | σ∗ (N7–O8) | 0.06 | 18.82 | 0.72 | 0.11 |
| LP (2) O11 | 1.9 | σ∗ (N10–O12) | 0.05 | 18.6 | 0.76 | 0.11 |
| LP (2) O8 | 1.9 | σ∗ (N7–O9) | 0.06 | 18.58 | 0.73 | 0.11 |
| π∗ (N7–O9) | 0.64 | π∗ (C4–C5) | 0.34 | 17.41 | 0.14 | 0.06 |
| π (C1–C3) | 1.72 | π∗ (C4–C5) | 0.34 | 15.35 | 0.29 | 0.06 |
| LP (2) O12 | 1.89 | σ∗ (C6–N10) | 0.1 | 13.93 | 0.58 | 0.08 |
| π (C4–C5) | 1.68 | π∗ (C2–C6) | 0.49 | 13.91 | 0.27 | 0.06 |
| LP (2) O9 | 1.9 | σ∗ (C4–N7) | 0.11 | 13.51 | 0.57 | 0.08 |
| LP (2) O8 | 1.9 | σ∗ (C4–N7) | 0.11 | 13.12 | 0.57 | 0.08 |
| π (N7–O9) | 1.99 | LP (3) O8 | 1.45 | 12.18 | 0.18 | 0.08 |
| π (C2–C6) | 1.59 | π∗ (C1–C3) | 0.24 | 11.19 | 0.31 | 0.06 |
E(2) is the energy of stabilization.
E(j)-E(i) is the energy difference between donor and acceptor i and j NBO orbitals.
F(i, j) is the Fock matrix element between i and j NBO orbitals.
5. Conclusion
Compound 3 is stabilized by C–H⋯π, N–O⋯π,N–H⋯O, N–H⋯N, C–H⋯N and C–H⋯O type interactions in crystalline phase. The interactions present in the molecule are unveiled by Hirshfeld surface analysis, RDG and topology analyses. The 3D energy framework calculation signifies that the dispersion energy has major contribution towards total interaction energy. The HOMO–LUMO energy gap of the molecule 3 is found to be 3.647 eV. The MEP map shows that the oxygen atoms present in the molecule exhibit electrophilic nature and hydrogen atoms are nucleophiles. This is substantiated by the Mulliken and natural atomic charge analyses with oxygen atoms having negative charges and hydrogen atoms having positive charges.
Declarations
Author contribution statement
Chandini K.M., Sridhar M.A.: Analyzed and interpreted the data; Wrote the paper.
Nagesh Khadri M. J.: Performed the experiments; Wrote the paper.
Amoghavarsha N: Performed the experiments.
Shaukath Ara Khanum: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data associated with this study has been deposited at Chemical Crystallographic Data Centre under the accession number 2151097.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e10047.
Acknowledgements
Nagesh Khadri M J is thankful to KSTePS, DST, Govt. of Karnataka for providing fellowship. Authors are grateful to the SAIF, IIT Madras for providing X-ray intensity data and IOE, Vijnana Bhavan, University of Mysuru for providing NMR data.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Laniyan T.A., Morakinyo O.M. Environmental sustainability and prevention of heavy metal pollution of some geo-materials within a city in southwestern Nigeria. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berhanu A.L., Gaurav, Mohiuddin I., Malik A.K., Aulakh J.S., Kumar V., Kim K.H. A review of the applications of Schiff bases as optical chemical sensors. Trends Anal. Chem. 2019;116:74–91. [Google Scholar]
- 3.Orojloo M., Amani S. Colorimetric detection of pollutant trivalent cations and HSO4− in aqueous media using a new Schiff-base probe: an experimental and DFT studies. Polycycl. Aromat. Comp. 2021;41:33–46. [Google Scholar]
- 4.Abu-Dief A.M., Mohamed I.M.A. A review on versatile applications of transition metal complexes incorporating Schiff bases. Beni-Suef Univ. J. Basic Appl. Sci. 2015;4:119–133. doi: 10.1016/j.bjbas.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uddin M.N., Siddique Z.A., Mase N., Uzzaman M., Shumi W. Oxotitanium(IV) complexes of some bis-unsymmetric Schiff bases: Synthesis, structural elucidation and biomedical applications. Appl. Organomet. Chem. 2019;33 [Google Scholar]
- 6.Ghosh P., Dey S.K., Ara M.H., Karim K., Islam A.B.M.N. A review on synthesis and versatile applications of some selected Schiff bases with their transition metal complexes, Egypt. J. Chem. 2019;62:523–547. [Google Scholar]
- 7.Manimaran A., Prabhakaran R., Deepa T., Natarajan K., Jayabalakrishnan C. Synthesis, spectral characterization, electrochemistry and catalytic activities of Cu(II) complexes of bifunctional tridentate Schiff bases containing O-N-O donors. Appl. Organomet. Chem. 2008;22:353–358. [Google Scholar]
- 8.Abdel-Shakour M., El-Said W.A., Abdellah I.M., Su R., El-Shafei A. Low-cost Schiff bases chromophores as efficient co-sensitizers for MH-13 in dye-sensitized solar cells. J. Mater. Sci. Mater. Electron. 2019;30:5081–5091. [Google Scholar]
- 9.Karman M., Romanowski G. Cis-dioxidomolybdenum(VI) complexes with chiral tetradentate Schiff bases: Synthesis, spectroscopic characterization and catalytic activity in sulfoxidation and epoxidation. Inorg. Chim. Acta. 2020;511 [Google Scholar]
- 10.Shetty P. Schiff bases: an overview of their corrosion inhibition activity in acid media against mild steel. Chem. Eng. Commun. 2020;207:985–1029. [Google Scholar]
- 11.Abdel-Rahman L.H., Abu-Dief A.M., Adam M.S.S., Hamdan S.K. Some new nano-sized mononuclear Cu(II) Schiff base complexes: design, characterization, molecular modeling and catalytic potentials in benzyl alcohol oxidation. Catal. Lett. 2016;146:1373–1396. [Google Scholar]
- 12.Nourifard F., Payehghadr M. Conductometric studies and application of new Schiff base ligand as carbon paste electrode modifier for mercury and cadmium determination. Int. J. Environ. Anal. Chem. 2016;96:552–567. [Google Scholar]
- 13.Singh L.P., Bhatnagar J.M. Copper(II) selective electrochemical sensor based on Schiff Base complexes. Talanta. 2004;64:313–319. doi: 10.1016/j.talanta.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Al Zoubi W., Al Mohanna N. Membrane sensors based on Schiff bases as chelating ionophores – a review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014;132:854–870. doi: 10.1016/j.saa.2014.04.176. [DOI] [PubMed] [Google Scholar]
- 15.Pillai R.R., Karrouchi K., Fettach S., Armaković S., Armaković S.J., Brik Y., Taoufik J., Radi S., El Abbes Faouzi M., Ansar M. Synthesis, spectroscopic characterization, reactive properties by DFT calculations, molecular dynamics simulations and biological evaluation of Schiff bases tethered 1,2,4-triazole and pyrazole rings. J. Mol. Struct. 2019;1177:47–54. [Google Scholar]
- 16.Bharti S.K., Nath G., Tilak R., Singh S.K. Synthesis, anti-bacterial and anti-fungal activities of some novel Schiff bases containing 2,4-disubstituted thiazole ring. Eur. J. Med. Chem. 2010;45:651–660. doi: 10.1016/j.ejmech.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Bushra Begum A., Rekha N.D., Vasantha Kumar B.C., Lakshmi Ranganatha V., Khanum S.A. Synthesis, characterization, biological and catalytic applications of transition metal complexes derived from Schiff base. Bioorg. Med. Chem. Lett. 2014;24:3559–3564. doi: 10.1016/j.bmcl.2014.05.046. [DOI] [PubMed] [Google Scholar]
- 18.Oiye É.N., Ribeiro M.F.M., Katayama J.M.T., Tadini M.C., Balbino M.A., Eleotério I.C., Magalhães J., Castro A.S., Silva R.S.M., da Cruz Júnior J.W., Dockal E.R., de Oliveira M.F. Electrochemical sensors containing Schiff bases and their transition metal complexes to detect analytes of forensic, pharmaceutical and environmental interest. A review. Crit. Rev. Anal. Chem. 2019;49:488–509. doi: 10.1080/10408347.2018.1561242. [DOI] [PubMed] [Google Scholar]
- 19.Abad N., Sallam H.H., Al-Ostoot F.H., Khamees H.A., Al-horaibi S.A., Khanum S.A., Ramli Y. Synthesis, crystal structure, DFT calculations, Hirshfeld surface analysis, energy frameworks, molecular dynamics and docking studies of novel isoxazolequinoxaline derivative (IZQ) as anti-cancer drug. J. Mol. Struct. 2021;1232 [Google Scholar]
- 20.SAINT-Plus B. Bruker AXS Inc.; Madison, Wisconsin, USA: 2004. SADABS (Version 2004/1. [Google Scholar]
- 21.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C: Struct. Chem. 2015;71(1):3–8. doi: 10.1107/S2053229614024218. .f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spek A.L. PLATON, an integrated tool for the analysis of the results of a single crystal structure determination. Acta Crystallogr. A: Found. Crystallogr. 1990;46(s1) c34-c34. [Google Scholar]
- 23.Macrae C.F., Bruno I.J., Chisholm J.A., Edgington P.R., McCabe P., Pidcock E., Wood P.A. Mercury CSD 2.0–new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41(2):466–470. [Google Scholar]
- 24.Jayatilaka D., Wolff S.K., Grimwood D.J., McKinnon J.J., Spackman M.A. CrystalExplorer: a tool for displaying Hirshfeld surfaces and visualising intermolecular interactions in molecular crystals. Acta Crysta. A-found. 2006;62 S90–S90. [Google Scholar]
- 25.McKinnon J.J., Jayatilaka D., Spackman M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007;37:3814–3816. doi: 10.1039/b704980c. [DOI] [PubMed] [Google Scholar]
- 26.Mackenzie C.F., Spackman P.R., Jayatilaka D., Spackman M.A. CrystalExplorer model energies and energy frameworks: extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ. 2017;4(5):575–587. doi: 10.1107/S205225251700848X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheese-man J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., IzmaylovJ A.F., Sonnenberg L., Williams-Young D., Ding F., Lip-parini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toy-ota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Keith T., Kobayashi R., Nor-mand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Mo-rokuma K., Farkas O., Foresman J.B., Fox D.J. Gaussian, Inc.; Walling-ford CT: 2016. Gaussian 09. [Google Scholar]
- 28.Dennington R., Keith T., Millam J. Semichem Inc.; Shawnee Mission, KS: 2019. GaussView, Version 6.1.1. [Google Scholar]
- 29.Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33(5):580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 30.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Tan S.L., Jotani M.M., Tiekink E.R. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. E: Crystallogr. Commun. 2019;75(3):308–318. doi: 10.1107/S2056989019001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spackman M.A., Jayatilaka D. Hirshfeld surface analysis. CrystEngComm. 2009;11:19–32. [Google Scholar]
- 33.Spackman M.A., McKinnon J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm. 2002;4(66):378–392. [Google Scholar]
- 34.Turner M.J., Thomas S.P., Shi M.W., Jayatilaka D., Spackman M.A. Energy frameworks: insights into interaction anisotropy and the mechanical properties of molecular crystals. Chem. Commun. 2015;51(18):3735–3738. doi: 10.1039/c4cc09074h. [DOI] [PubMed] [Google Scholar]
- 35.Al-shawi J.M., Hussain K.A., Ismael S.M. Investigation of molecular structure, chemical reactivity and stability for pyrrole substitutes (dual anchoring system) using density functional theory. J. Basrah Researches. 2015;(3):41. Sciences. [Google Scholar]
- 36.Costa R.A., Pitt P.O., Pinheiro M.L.B., Oliveira K.M., Salome K.S., Barison A., Costa E.V. Spectroscopic investigation, vibrational assignments, HOMO-LUMO, NBO, MEP analysis and molecular docking studies of oxoaporphine alkaloid liriodenine. Spectrochim. Acta Mol. Biomol. Spectrosc. 2017;174:94–104. doi: 10.1016/j.saa.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Govindasamy P., Gunasekaran S., Srinivasan S. Molecular geometry, conformational, vibrational spectroscopic, molecular orbital and Mulliken charge analysis of 2-acetoxybenzoic acid. Spectrochim. Acta Mol. Biomol. Spectrosc. 2014;130:329–336. doi: 10.1016/j.saa.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Wu J., Zhu J., Wang Z., Zhou J., Cen K. Removal of oxygen functional groups in lignite by hydrothermal dewatering: an experimental and DFT study. Fuel. 2016;178:85–92. [Google Scholar]
- 39.Prabhaharan M., Prabakaran A.R., Gunasekaran S., Srinivasan S. DFT studies on vibrational spectra, HOMO–LUMO, NBO and thermodynamic function analysis of cyanuric fluoride. Spectrochim. Acta Mol. Biomol. Spectrosc. 2015;136:494–503. doi: 10.1016/j.saa.2014.09.062. [DOI] [PubMed] [Google Scholar]
- 40.Shukla S., Srivastava A., Kumar P., Tandon P., Maurya R., Singh R.B. Vibrational spectroscopic, NBO, AIM, and multiwfn study of tectorigenin: a DFT approach. J. Mol. Struct. 2020;1217 [Google Scholar]
- 41.Johnson E.R., Keinan S., Mori-Sánchez P., Contreras-García J., Cohen A.J., Yang W. Revealing noncovalent interactions. J. Am. Chem. Soc. 2010;132(18):6498–6506. doi: 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kavimani M., Balachandran V., Narayana B., Vanasundari K., Revathi B. Topological analysis (BCP) of vibrational spectroscopic studies, docking, RDG, DSSC, Fukui functions and chemical reactivity of 2-methylphenylacetic acid. Spectrochim. Acta Mol. Biomol. Spectrosc. 2018;190:47–60. doi: 10.1016/j.saa.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Mıhçıokur Ö., Özpozan T. Molecular structure, vibrational spectroscopic analysis (IR & Raman), HOMO-LUMO and NBO analysis of anti-cancer drug sunitinib using DFT method. J. Mol. Struct. 2017;1149:27–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at Chemical Crystallographic Data Centre under the accession number 2151097.