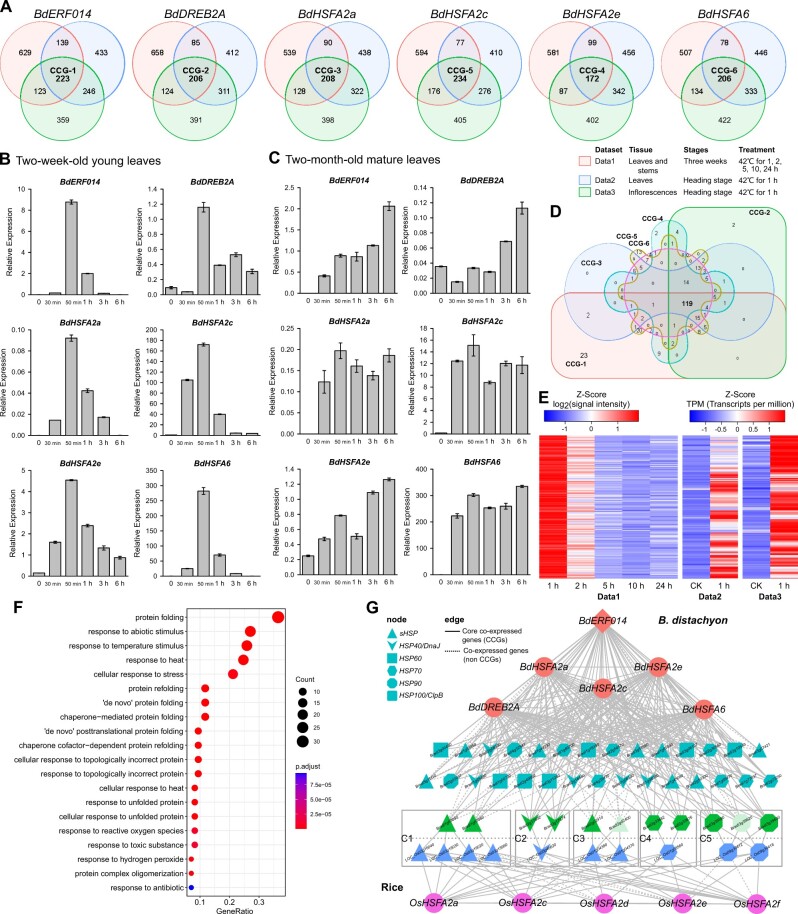
Figure 6.
BdERF014 might engage in HSR by cross-talking with preexisting HSFAs-HSPs signaling pathways. A, Venn diagram showing overlap of the genes highly co-expressed with BdERF014, BdDREB2A, BdHSFA2a, BdHSFA2c, BdHSFA2e, BdHSFA6, respectively, under heat stress conditions. The numbers of genes in each region of the diagram are indicated. B and C, The expression analysis of BdERF014 and five typical heat-stress responsive TF genes under heat stress. After treated at 42°C, the leaves of the 2-week-old seedlings (B) and the two-month-old plants (C) were harvested for the expression analysis. β-Actin gene was used as the internal control for normalization. The relative expression levels were calculated by the 2−ΔCt method. The error bars indicate the SE of three independent biological replicates. D, Venn diagram showing overlap of the CCGs identified in (A). E, Heatmap revealing the expression of CCGs of BdERF014. F, The top 20 GO terms enriched in CCGs of BdERF014. G, Co-expression network of the CCGs of heat stress related TF genes in B. distachyon and rice. Diamond and circular nodes indicate TF genes of B. distachyon and rice. Nodes with other shapes correspond to the groups in the legend. Five rectangular boxes indicate the five HSP gene clusters, C1–C5, respectively. Brachypodium distachyon genes are above the dotted line and rice genes are below. Edge types correspond to the edge information in the legend.