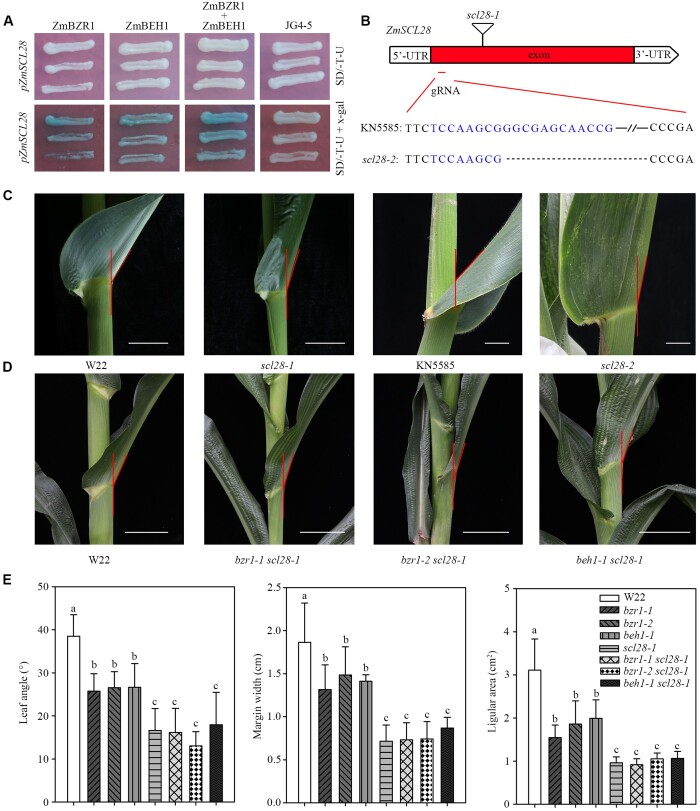
Figure 7.
ZmSCL28 works downstream of ZmBZR1 and ZmBEH1. A, Y1H assays showed that ZmBZR1 and ZmBEH1 bound to the promoter of ZmSCL28. B, Gene model presents the positions of two mutant alleles in ZmSCL28. The triangle shows the position of a Mu insertion (UFMu-09491) from line scl28-1 in the W22 genetic background. The position of a single-guide RNA (sgRNA) designed to mutate ZmSCL28 using CRISPR/Cas9 technology is labled in exon. The sequence of sgRNA is highlighted, and the symbols “…” indicate the deletion caused by CRISPR/Cas9-induced mutations in line scl28-2 in the KN5585 genetic background. C, Leaf angle morphology in W22 and scl28-1, KN5585 and scl28-2 at the V10 stage. Bar, 5 cm. D, Leaf angle morphology in the WT W22 and double mutants of bzr1-1 scl28-1, bzr1-2 scl28-1, and beh1-1 scl28-1 at the V10 stage. Bar, 5 cm. E, Quantitative measurements of leaf angle (n = 20), auricle margin width (n = 10), and auricle area (n = 10) in single and double mutants. Error bars are SD. Different letters above the columns indicate statistically significant differences between groups (Student’s t test).