Extended Data Fig. 7. Flow diagram of the fecal microbiome sample collection.
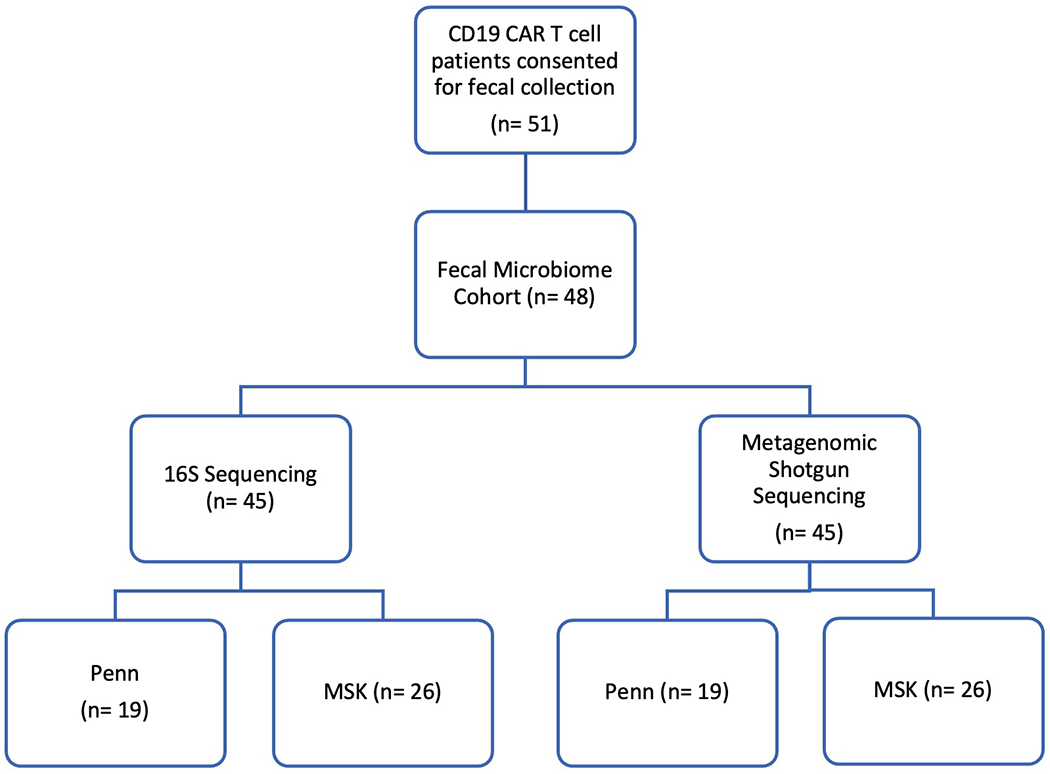
Fifty-one unique patients were collected upon informed consent. Of the fifty-one patients, one patient did not have sufficient fecal material for sequencing and two patients failed during the amplification or quality control step. Following these exclusions, there were forty-eight patients in the fecal microbiome cohort. Of these patients, we successfully amplified and sequenced the 16S ribosomal RNA gene with ≥ 200 reads per sample from forty-five patients. Forty-five patients passed quality control measures for metagenomic shotgun sequencing. There were three non-overlapping patients in the 16S and shotgun sequencing cohorts. Hence, there were 48 unique patients in the fecal microbiome cohort.
