Figure 1.
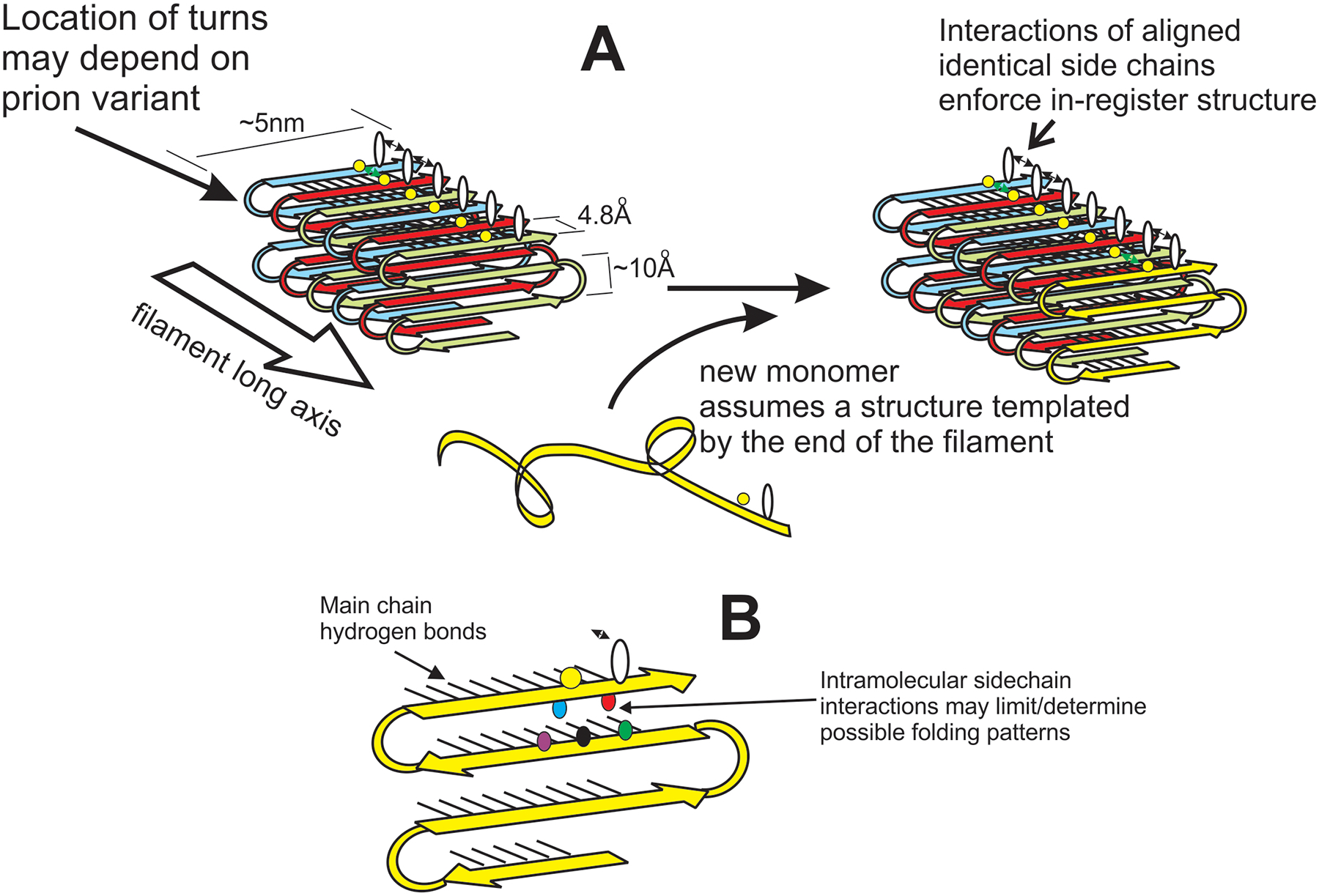
The in-register parallel architecture of yeast prion amyloid can explain templating of conformation. A. The unstructured prion domain of molecules newly joining the end of the amyloid filament are directed to assume the same conformation as molecules already in the filament by the favorable interactions between identical side chains that can only occur if the molecules are in-register [77]. B. Three interactions of an amino acid residue are shown. The black lines show the main chain hydrogen bonds of the β-sheet. The black double-headed arrow indicates the intermolecular side-chain interactions between identical residues (emphasized in A). Also shown are the interactions between non-identical side chains in the plane perpendicular to the long axis of the filaments. These interactions have been studied by X-ray crystallography of short peptides [78].
