Fig. 3.
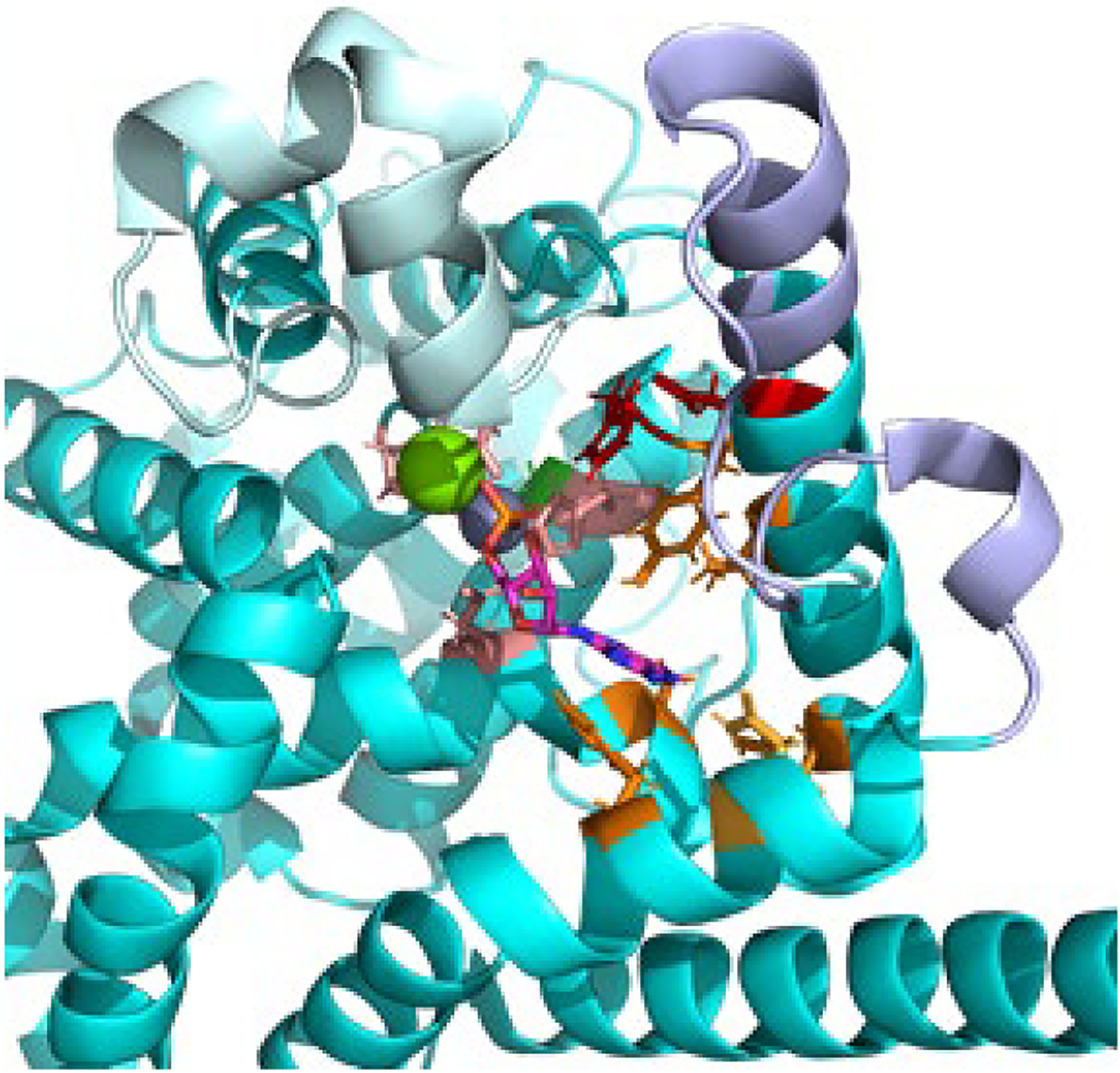
Catalytic domain of PDE6 The catalytic domain of Pβ (PDBID: 6mzb, cyan) was aligned with the PDE5 catalytic domain crystal structure (PDBID: 1t9s) in order to visualize the divalent cations Zn (gray) and Mg (green) and the 5’-GMP product in the enzyme active site. The side chains of residues involved in the metal binding (M-site, light brown), the nucleotide binding site (Q-site, orange), or participating in the catalytic reaction mechanism (red) are shown. The residue implicated in catalytic acceleration (Gly562, green [21]) is located behind the Zn atom. Also shown are the conformationally dynamic H-loop (pale cyan) and M-loop (lavender) implicated in regulation of the catalytic rate.
