Abstract
Purpose
To investigate the ocular surface microbiome of patients with unilateral or asymmetric glaucoma being treated with topical ophthalmic medications in one eye and to determine whether microbial community changes were related to measures of ocular surface disease.
Methods
V3-V4 16S rRNA sequencing was conducted on ocular surface swabs collected from both eyes of 17 subjects: 10 patients with asymmetric/unilateral glaucoma using topical glaucoma therapy on only one eye and seven age-matched, healthy controls with no history of ocular disease or eyedrop use. Samples were categorized into three groups: patients’ glaucomatous eye treated with eyedrops, patients’ contralateral eye without eyedrops, and healthy control eyes. Comparisons were made for microbial diversity and composition, with differences in composition tested for association with ocular surface disease measures including tear meniscus height, tear break-up time, and Dry Eye Questionnaire.
Results
Samples obtained from the patients’ treated and untreated eyes both had significantly greater alpha-diversity and relative abundance of gram-negative organisms compared to healthy controls. The microbial composition of patient eyes was associated with decreased tear meniscus height and tear break-up time, whereas metagenomic predictions, based on 16S rRNA data, suggested increased synthesis of lipopolysaccharide.
Conclusions
The ocular surface microbiome of patients taking unilateral preserved glaucoma drops is characterized by a highly diverse array of gram-negative bacteria that is significantly different from the predominantly gram-positive microbes detected on healthy control eyes. These compositional differences were associated with decreased tear film measures and distinct inferred protein synthesis pathways, suggesting a potential link between microbial alterations and ocular surface inflammation.
Keywords: microbiome, ocular surface, preservatives, glaucoma medications, glaucoma
Glaucoma is a chronic progressive optic neuropathy involving retinal ganglion cells and is the second leading cause of blindness in the world.1 There are multifactorial mechanisms responsible for disease progression including intraocular pressure, vascular perfusion,2 chronic inflammation,3 and oxidative stress.4 Treatment modalities target the reduction of intraocular pressure using topical eyedrop medications, laser trabeculoplasty, or filtering surgery. When patients are started on pressure-reducing eyedrops, the expectation is that they will continue using these medications for the rest of their lives, with many patients on several different eyedrops for adequate pressure control. Patient compliance with daily application of eyedrops is important to preventing poor visual outcomes.
Ocular discomfort associated with use of glaucoma eyedrops may adversely affect compliance. Large cohort studies have shown that glaucoma eyedrops containing the preservative benzalkonium chloride (BAK) increases the frequency of ocular irritative symptoms including burning, dry eye sensation, and tearing,5,6 with more severe symptoms reported by patients on greater than one eyedrop7 and improvement of symptoms when switching to preservative-free formulations.8 The use of preservatives in eyedrops have been associated with decreased tear break-up times, tear film instability,9 decreased tear turnover,10 and increased corneal epithelial permeability,11 while causing structural changes such as decreased goblet cell density and mucus secretion12 that create an ocular surface environment similar to that of dry eye disease. Prior studies have also investigated the effects of the preservative on increasing inflammatory cell populations,13 conjunctival collagen thickness,14 and corneal cell death.15
The primary role of BAK is to prevent the growth of pathogenic bacteria in medication bottles by acting as a detergent to disrupt bacterial cell walls and release cytoplasmic contents.9 We hypothesize that BAK has similar antimicrobial effects on organisms that colonize the ocular surface and may cause significant alterations in the types of organisms (“microbiota”) on the ocular surface. There is evidence to suggest that the healthy ocular surface harbors a stable composition of organisms16-18 that, when disrupted, can predispose patients to developing ocular surface infection or inflammation.19,20 In this study, we sequenced and analyzed the ocular surface microbiota of both eyes in a cohort of patients with asymmetric or unilateral glaucoma, receiving eyedrop treatment in only one eye, to determine whether the use of preserved glaucoma eyedrops is associated with alterations of the ocular surface microbiota and to determine whether these changes are associated with measures of ocular surface disease. The untreated eyes in our cohort were used as internal controls; however, we hypothesized that topical glaucoma drops may also affect the microbiome of the untreated eyes and so included a cohort of age-matched subjects who were not taking any eye drops as a secondary control population.
Methods
Study Design
This case-control study was approved by the Columbia University Medical Center Human Subjects Division institutional review board (Protocol no. AAAR6267). Research adhered to the tenets of the Declaration of Helsinki and was conducted in accordance with the Health Insurance Portability and Accountability Act regulations. Written informed consent was obtained from all subjects. Participants were screened and enrolled from the same Columbia Doctors clinical ophthalmology offices that housed multiple affiliated glaucoma specialists. The study had 17 participants, categorized into 10 patients and seven healthy controls. Patients were selected if they were diagnosed with any type of glaucoma and were currently using preserved topical glaucoma therapy for over one month duration in only one eye. Healthy controls were age-matched volunteer participants without any history of ocular disease or eyedrop use. Exclusion criteria included subjects under the age of 18 years, history of systemic or topical antibiotics or corticosteroid use within the past three months, history of incisional glaucoma surgery (including trabeculectomy and glaucoma drainage implants), and any ocular surgical history within the past three months.
Sample Collection
Ocular swab samples were obtained from each eye at both the conjunctiva and eyelid margin, with a total of four samples collected for each subject. Before sample collection, a drop of proparacaine from a previously unopened bottle was administered to each eye. The conjunctival and eyelid margin microbiomes were collected separately from eye each using a sterile swab (Isohelix Buccal DNA/RNA Swab; Cell Projects Ltd., Kent, UK) swept three times over the inferior conjunctival fornix and lower eyelid margin, respectively. A total of four samples were collected from each subject. Additional air samples were collected with a sterile swab exposed to the surrounding environment and a drop of proparacaine solution—these were used as negative controls during each sample collection event. Samples were then placed into pyrogen-free 1.5 mL cryovials (Thermo Scientific) and immediately frozen at -80°C. Ocular samples were categorized into three groups: samples collected from the patients’ glaucomatous eye receiving topical medication (“Eyedrops”), samples collected from the patients’ contralateral eye without treatment (“No eyedrops”), and samples collected from healthy control eyes (“Controls”).
DNA Extraction and Sequencing
DNA extraction was performed using the Allprep PowerViral DNA/RNA Kit (Qiagen, Venlo, the Netherlands) with modifications. Briefly, swabs were suspended in 100 μL PowerBead solution and spun in a centrifuge at 24,000 RPM for five minutes. Swabs were then removed and the resulting eluant was supplemented with an additional 500 μL of PowerBead solution. The remainder of protocol was carried out per the manufacturers’ protocol. All samples produced low DNA yields (<2 ng/uL) as measured by Quant-iT dsDNA BR assay (Invitrogen, Waltham, MA, USA). Library preparation and sequencing was attempted on all samples with experimental and corresponding negative control samples processed concurrently.
The V3-V4 hypervariable region of the 16S rRNA gene was amplified using previously described primers and protocol.21 Concentration and purity of PCR products were determined by PicoGreen (Invitrogen, Carlsbad, CA, USA) and an Agilent Bioanalyzer 2100 DNA 1000 chip (Agilent Technologies, Santa Clara, CA), respectively. Amplicons were pooled in equimolar concentrations and sequencing performed on the Illumina MiSeq and the MiSeq reagent kit v3 (600-cycle) according to the manufacturers’ instructions.
Clinical Examination
Study participants were evaluated for ocular surface disease using the non-invasive OCULUS Keratograph 5M (OCULUS, Inc., Arlington, WA, USA). Clinical measurements included the OCULUS noninvasive keratograph tear break-up time (NIKBUT), the OCULUS tear meniscus height, and the RPS InflammaDry Detector (Quidel, San Diego, CA, USA), a noninvasive assay that detects the presence of matrix metalloproteinase-9.22 Participants also completed a survey that included the validated five-item Dry Eye Questionnaire (DEQ-5)23 along with questions related to ocular history of eye disease and associated topical therapies. Additional qualitative information regarding disease course and severity were captured using the participants’ medical records.
Statistical Analysis
Demographics (age and sex), ocular history (eye discomfort rating, dry eye symptoms, frequency of watery eyes, DEQ-5 score), RPS result, tear meniscus height, tear meniscus height score, first tear break-up time, average tear break-up time, and NIKBUT scores were compared between eye samples treated with eyedrops, no eyedrops, and healthy controls. Differences in continuous variables were evaluated using Wilcoxon rank sum or Kruskal-Wallis test, where appropriate. Comparison of categorical variables were conducted using Fisher's exact test.
The raw 16S rRNA sequencing reads were pre-processed using the DADA2 pipeline package24 in QIIME2 (version 2020.6)25 to remove primers, demultiplex sequences, filter reads for quality (Q25 cutoff), and remove chimeric sequences. The output amplicon sequence variant (ASV) table was generated for ocular swab samples and air swab samples. The QIIME2 quality-control package was used for removal of contaminants by excluding ASVs present in 11 of 12 air swab control samples from the experimental samples, containing a range of 10 to 2083 feature counts (Supplemental Fig. S1). A single air swab sample (AIR-6267-CTL005) produced a substantially higher sequence read count (38,275) than that of all other control swab samples and a bacterial profile composition like that of the secondary healthy control subjects. Thus we considered that this single control likely consisted of experimentally relevant features that were not representative of other air swabs (Supplemental Data S1); we thus did not remove these data from our dataset. Taxonomic classification of ASVs using the SILVA v138 database26 and diversity analyses (alpha and beta diversity) tested against sample metadata factors were performed using QIIME2. Alpha- and beta-diversity metrics were rarefied to a sequencing depth of 1000 reads per sample, with no significant changes in diversity measures with increased depth confirmed by alpha rarefaction curves. For our analysis, 24 of 28 healthy control samples (margin and conjunctiva of each eye of seven subjects), 16 of 20 patient treated eye samples, and 15 of 20 patient untreated eye samples had a read depth above this threshold.
Alpha-diversity metrics (Shannon's and Simpson's index) and beta-diversity metrics (Bray-Curtis Dissimilarity, Jaccard's index, weighted Unifrac, and Unweighted Unifrac distance matrices) were calculated using the QIIME2 q2-diversity plug-in.27 Alpha-diversity metrics were compared using Kruskal-Wallis test with FDR-correction for multiple comparisons. Beta-diversity metrics were compared and tested for associations with clinical data using permutational multivariate analysis of variance (PERMANOVA) tests with 999 permutations. In our results, we present Shannon's diversity to represent alpha-diversity and Bray-Curtis for beta-diversity, with additional diversity metrics used to validate our findings. Principal coordinate analysis plots were generated using the QIIME2 emperor plugin28,29 to visualize findings. Correlation of alpha-diversity metrics to continuous variables were performed using Spearman's rank correlation. Linear discriminant analysis coupled with effect size measurements algorithm30 was used to perform nonparametric Kruskal-Wallis tests and identify bacterial taxa whose relative abundance was significantly different in a group of interest compared to controls. To compare the differential abundance of ASVs between groups, two separate models were conducted using a negative binomial distribution in DESeq231 and a centered log-ratio transformation of compositional data. The two models did not have significant differences in results, and we presented findings using DESeq2. Significant associations of ASVs to continuous outcome variables (e.g., tear meniscus height) were also conducted using DESeq2. Metagenome inference was performed using PiCRUST232 with statistical analysis of relatively abundant pathways using ALDEx2.33 Statistical significance was defined as Benjamini-Hochberg FDR corrected P values < 0.05, q value < 0.05, and linear discriminant analysis score > 2. The statistical analyses were performed using built-in QIIME2 packages (version 2020.6) and R (version 4.0.2).
Samples from patients and from healthy controls were sequenced on two separate runs. To ensure there was no significant batch effect, the air swab controls were compared across sequencing runs. There was no significant difference in the PERMANOVA of the microbial composition between runs, P = 0.359 (Supplemental Fig. S1).
In our preliminary analysis, we stratified comparisons for sampling site (conjunctiva vs. margin) for all subjects and sampled eye of health controls (left vs. right). We found no differences in measures of alpha- and beta-diversity and did not identify differentially abundant ASVs based on sampled site and eye (Supplemental Figs. S2, S3), suggesting that these factors were unlikely to be independent confounders. Here we present results where samples from different sampled site (patients and healthy controls) and eye (healthy controls only) were analyzed together to minimize noise and artefact associated with smaller sample sizes and low burden microbiome samples. Statistical analyses were adjusted for multiple comparisons by adopting a mixed effects model, treating subject ID as a random effect.
Results
Demographics and Clinical Characteristics
A total of 17 subjects participated in this study, 10 patients with a diagnosis of asymmetric/unilateral glaucoma and seven control subjects without any history of ocular disease (Table 1). The average age of patients was 71 years old (range 32–91), and average age for control subjects was 66.9 years (range 33–83). Of the 10 patients, six (60%) carried a diagnosis of glaucoma for more than two years, whereas the other four (40%) were diagnosed within one year prior to the study. Types of asymmetric/unilateral glaucoma included pseudoexfoliative (n = 5), primary open angle (n = 2), chronic angle closure (n = 1), traumatic (n = 1), and ocular hypertension (n = 1). Five of the patients had used topical glaucoma therapy for two to five years, whereas the remainder had used these eyedrops for less than one year. In addition, five of the patients used one glaucoma topical therapy drop, which included prostaglandin (n = 1), beta blocker (n = 2), carbonic anhydrase inhibitor (n = 1), and combination beta blocker/carbonic anhydrase inhibitor (n = 1) whereas the other five used two or more eyedrops. Five patients reported daily use of eyedrop medications, and five others reported twice or more daily use. All topical glaucoma medications used by patients during the study contained the preservative BAK. Duration, frequency, and type of glaucoma therapy were not associated with differences in ocular bacterial diversity or microbiome composition.
Table 1.
Demographics and Ocular History of Patients With Glaucoma in One Eye Receiving Eyedrops, No Glaucoma in the Opposite Eye With No Eyedrops, and Control Subjects Without History of Glaucoma
| Unilateral Glaucoma Patients (n = 10 Patients) | ||||||
|---|---|---|---|---|---|---|
| Eyedrops (E) | No Drops (ND) | Controls (C) | ||||
| n = 20 | P Value (E-C) | N = 20 | P Value (E-ND) | N = 28 | P Value (ND-C) | |
| Demographics | ||||||
| Sex | ||||||
| Female | 6 (30%) | 0.08 | 6 (30%) | — | 16 (57.1%) | 0.08 |
| Male | 14 (70%) | 0.08 | 14 (70%) | — | 12 (42.9%) | 0.08 |
| Age, years | 71 | 0.40 | 71 | — | 66.9 | 0.40 |
| Ocular History | ||||||
| Eye discomfort | ||||||
| Frequency | 1.10 | 0.26 | 0.40 | 0.08 | 0.57 | 0.25 |
| Intensity | 1.60 | 0.04 | 0.35 | 0.04 | 0.57 | 0.48 |
| Dry eye symptoms | ||||||
| Frequency | 0.60 | 0.07 | 0.30 | 0.39 | 1.00 | 0.004 |
| Intensity | 0.70 | 0.22 | 0.30 | 0.32 | 1.07 | 0.02 |
| Frequency of watery eyes | 0.90 | 0.92 | 0.40 | 0.32 | 0.57 | 0.23 |
| DEQ-5 Score | 4.90 | 0.77 | 1.75 | 0.03 | 3.79 | 0.003 |
| Eyedrop usage | ||||||
| Frequency | 3.50 | — | — | — | — | — |
| Duration | 1.10 | — | — | — | — | — |
| RPS result | ||||||
| Positive | 8 (40%) | 0.04 | 6 (30%) | 0.74 | 20 (71.4%) | 0.008 |
| Negative | 12 (60%) | 0.04 | 14 (70%) | 0.74 | 8 (28.6%) | 0.008 |
| Tear meniscus height (in mm) | 0.408 | <0.001 | 0.557 | 0.03 | 0.548 | 0.28 |
| Tear meniscus height score | 2.400 | <0.001 | 2.700 | 0.29 | 2.714 | 0.003 |
| First tear break-up time (sec) | 7.001 | 0.009 | 10.011 | 0.004 | 9.558 | 0.29 |
| Average tear break-up time (sec) | 13.989 | 0.47 | 16.338 | 0.51 | 15.651 | 0.36 |
| NIKBUT score | 3.444 | 0.06 | 3.375 | 0.37 | 3.417 | 0.01 |
Numerical values are averages calculated from “n” number of samples. Two ocular swab samples were obtained from each eye at the conjunctiva and eyelid margin, for a total of four samples collected for each individual patient (10 patients = 40 patient samples) and control subject (seven controls = 28 control samples). Percentage values are calculated as number of samples divided by the group total. P values are displayed for pairwise comparisons of the two columns to the left preceding the value (E-C = eyedrops vs. controls; E-ND = eyedrops vs. no drops; ND-C = no drops vs. controls). P values are calculated using Wilcoxon Rank Sum or Kruskal-Wallis test. Eye discomfort, dry eye symptoms, and frequency of watery eyes are numerical ratings that are added up to obtain the DEQ-5 score. DEQ-5 with frequency of eye discomfort, dryness, and watery eyes rated on a 0-4 scale (never to constantly) and intensity of eye discomfort and dryness rated on a 0-5 scale (never to very intense).
NIKBUT score (by tear break-up time in seconds): <7 seconds = 1; 7–11 seconds = 2; 11–15 seconds = 3; >15 seconds = 4.
The average total score of the DEQ-5 was highest for the patients’ glaucomatous eyes receiving drops (4.90), followed by healthy control subject eyes (3.79), which were both significantly higher than the score for patients’ eyes without drops (1.75, Kruskal-Wallis P = 0.03 and P = 0.003, respectively). Patient eyes receiving topical therapy had a significantly lower tear meniscus height, tear meniscus height score, and first tear break-up time compared to patient eyes without drops and healthy control eyes (Table 1). A significantly higher proportion of healthy control subjects had a positive RPS result (71.4%) compared to patient eyes receiving topical therapy (40%) and without topical therapy (30%).
Ocular Surface Alpha Diversity is Increased in Patients Using Topical Glaucoma Medications
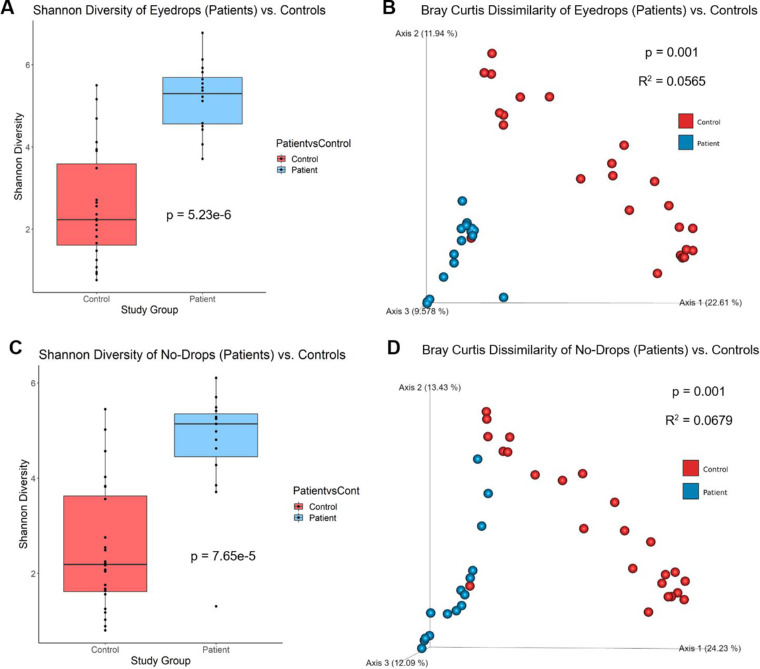
Measures of alpha diversity, which summarizes bacterial types (richness) and their distribution (evenness) using a single integer, and beta diversity (microbiota composition) were analyzed. Shannon diversity was significantly increased in the eyes of patients receiving drops (n = 16 samples) compared to healthy controls (n = 24 samples), an observation that was confirmed when Simpson's diversity was also compared (P < 0.0001, Kruskal Wallis) (Fig. 1A). Patient eye samples without drops (n = 15 samples) also demonstrated increased Shannon diversity compared to healthy controls (n = 24 samples) and was similarly validated by Simpson's (P < 0.0001, Kruskal-Wallis) (Fig. 1C). There were no significant differences in Shannon diversity (P = 0.36, Kruskal-Wallis) between the patient glaucomatous treated eye (n = 16) and the contralateral non-treated eye (n = 15) (Fig. 2A). Alpha-diversity indices had no significant correlation with DEQ-5 score (r = 0.16, −0.11, P = 0.27, 0.16), tear meniscus height (r = −0.19, −0.063, P = 0.20, 0.67), first tear break-up time (r = 0.21, 0.016, P = 0.22, 0.92), or average tear break-up time (r = −0.041, −0.077, P = 0.81, 0.64) with Spearman coefficients and P values listed for Shannon diversity measures from patient eye samples receiving drops and with no drops, respectively.
Figure 1.
Alpha and Beta Diversity for patient eyes treated with eyedrops and no-drops compared to healthy control eyes. (A) Patient eyes treated with eyedrops (n = 16) exhibited significantly greater alpha-diversity measures compared to healthy control eyes (n = 24) in Shannon diversity (P = 5.23e-6, Kruskal-Wallis). There were also significant differences in microbial composition, demonstrated by principal coordinate analysis of beta diversity, between patient eye samples with eyedrops compared to controls. (B) Differences were seen in beta-diversity based on Bray Curtis distances (P = 0.001, R2 = 0.0565, PERMANOVA). (C) Patient eyes without eyedrops (n = 15) also exhibited significantly greater alpha-diversity measures compared to healthy controls (n = 24) in Shannon diversity (P = 7.65e-5, Kruskal-Wallis). There were significant differences in microbial composition between patient eye samples without eyedrops compared to controls. (D) Differences were seen in beta-diversity based on Bray Curtis distances (P = 0.001, R2 = 0.0679, PERMANOVA). The axis values in the beta-diversity plot are the percentage of variance of phylogenetic beta diversity.
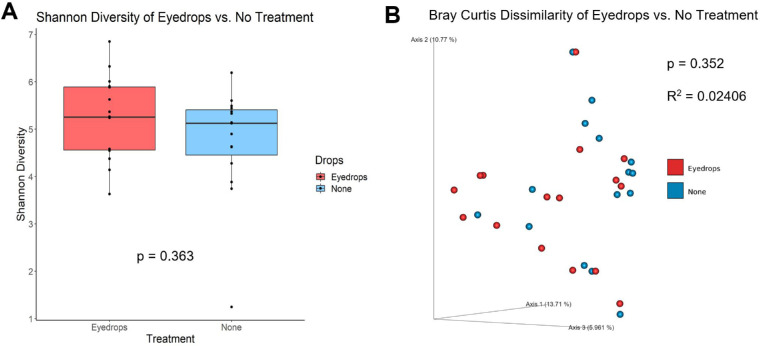
Figure 2.
Alpha and Beta Diversity for patient eyes treated with eyedrops (“Eyedrops”) and contralateral eyes without eyedrops (“No Treatment”). Patients were defined as study participants with a diagnosis of unilateral glaucoma, receiving eyedrop treatment in the glaucomatous eye (“eyedrops,” treated) but not the contralateral eye (“no treatment,” untreated). (A) There were no differences in alpha-diversity measures between the treated (n = 16) and untreated eye samples (n = 15) in Shannon diversity (P = 0.363, Kruskal-Wallis). There were no significant differences in microbiome composition between patient eyes treated with drops compared with untreated eyes, as no differences were seen in beta-diversity based on Bray Curtis distances (P = 0.352, R2 = 0.02406, PERMANOVA). The axis values in beta-diversity plots are the percentage of variance of phylogenetic beta diversity.
The Ocular Surface Microbial Composition and Abundance of Patients Using Topical Glaucoma Medications is Distinct From Those of Healthy Controls
The Bray-Curtis dissimilarity (beta diversity) showed a significant difference in microbial composition between both patient eyes receiving drops and without drops compared to healthy controls (P = 0.001, PERMANOVA), with no difference between the treated patient eye and the contralateral untreated patient eye (Figs. 1B, 1D, 2B; Table 2). These findings were confirmed by weighted and unweighted Unifrac distances and Jaccard Index. When adjusting for treatment status, patient versus control status, age, and sex, the differences in microbiota composition observed between patient and control eye samples were significantly associated with frequency of watery eyes, tear meniscus height, first tear break-up time, and average tear break-up time (Table 3).
Table 2.
Beta-Diversity Analysis Results for Comparisons Based on Sampling Eye and Location, Presence/Absence of Eyedrops, and Presence/Absence of Glaucoma
| P Values | |||
|---|---|---|---|
| Comparisons | Bray-Curtis Dissimilarity | Jaccard Distance Matrix | Weighted Unifrac Distance |
| Right vs. left eye | |||
| Control Conj. (n = 7 for both groups) | 0.942 | 0.704 | 0.116 |
| Control Margin (n = 7 for both groups) | 0.501 | 0.987 | 0.326 |
| Conj. vs. Margin | |||
| Control subjects (n = 14 for both groups) | 0.978 | 0.022 | 0.002 |
| Treated patients (n = 10 for both groups) | 0.383 | 0.155 | 0.209 |
| Drops vs. Controls (n = 20 vs. n = 28) | 0.001 | 0.001 | 0.001 |
| No drops vs. Controls (n = 20 vs. n = 28) | 0.001 | 0.001 | 0.001 |
| Drops vs. No drops (n = 20 for both groups) | 0.352 | 0.583 | 0.614 |
| Untreated glaucoma eyes vs. no glaucoma eyes (n = 4 vs. n = 16) | 0.850 | 0.975 | 0.795 |
Conj., conjunctiva; Drops, patient eye treated with eyedrops; No drops, patient eye without eyedrops; Control, study subjects without glaucoma.
n = number of samples for each comparison group in the order listed; the number of data points that appear on corresponding principal coordinates analysis (PCoA) plots may be less than listed here if some samples contained a lower feature count than the rarefaction depth. P values displayed were calculated by PERMANOVA.
Table 3.
Association Between Ocular Symptom Indexes With Beta Diversity Metric (Bray-Curtis Distances)
| PERMANOVA* | ||
|---|---|---|
| Ocular Symptom Measurements | R 2 | P Value |
| Eyedrops vs Control | 0.0565 | 0.001 |
| Eye discomfort | ||
| Frequency | 0.0195 | 0.542 |
| Intensity | 0.0142 | 0.959 |
| Dry eye symptoms | ||
| Frequency | 0.0206 | 0.412 |
| Intensity | 0.0233 | 0.226 |
| Frequency of watery eyes | 0.0359 | 0.010 |
| DEQ-5 Score | 0.0250 | 0.153 |
| Eyedrop usage | ||
| Frequency | 0.0197 | 0.546 |
| Duration | 0.0201 | 0.516 |
| RPS result | 0.0178 | 0.717 |
| Tear meniscus height | 0.0234 | 0.202 |
| Tear meniscus height score | 0.0174 | 0.781 |
| First tear break-up time | 0.0248 | 0.079 |
| Average tear break-up time | 0.0253 | 0.071 |
| NIKBUT score | 0.0452 | 0.145 |
| No Drops vs Control | 0.0679 | 0.001 |
| Eye discomfort | ||
| Frequency | 0.0204 | 0.447 |
| Intensity | 0.0179 | 0.686 |
| Dry eye symptoms | ||
| Frequency | 0.0223 | 0.306 |
| Intensity | 0.0223 | 0.253 |
| Frequency of watery eyes | 0.0379 | 0.004 |
| DEQ-5 Score | 0.0230 | 0.216 |
| Eyedrop usage | ||
| Frequency | 0.0248 | 0.151 |
| Duration | 0.0211 | 0.387 |
| RPS result | 0.0245 | 0.137 |
| Tear meniscus height | 0.0305 | 0.036 |
| Tear meniscus height score | 0.0261 | 0.127 |
| First tear break-up time | 0.0288 | 0.019 |
| Average tear break-up time | 0.0315 | 0.015 |
| NIKBUT score | 0.0495 | 0.052 |
Adjusted for treatment status, patient vs control status, age, and sex.
Note: PERMANOVA test results for association shown for eyedrops vs. control and no-drops vs. control comparisons. Eyedrops = patient eye samples treated with eyedrops; No Drops = patient eye samples without eyedrops; Controls = healthy control subjects without glaucoma.
RPS Result = RPS InflammaDry Detector test.
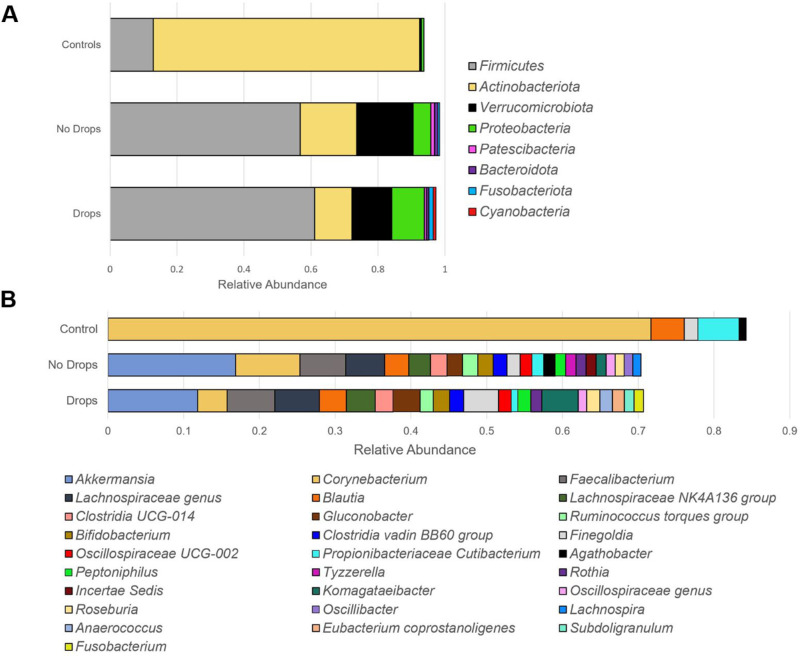
The ocular surface microbiota obtained from healthy controls was dominated by Actinobacteriota (79.5% of taxa) and Firmicutes (12.9%) at the phylum level, with Corynebacterium (71.7%), Cutibacterium (5.4%), and Blautia (4.4%) being the most common classifications at the genus level. In comparison, samples obtained from patient glaucomatous eyes treated with drops were primarily composed of Firmicutes (61.1%) and Verrucomicrobiota (11.8%) with decreased abundance of Actinobacteriota (11.2%), while Akkermansia (11.8%), Faecalibacterium (6.3%), Lachnospiraceae (5.9%), Komagataeibacter (4.8%), Finegoldia (4.6%), Corynebacterium (3.9%), and Blautia (3.6%) were the most common classifications at the genus level. Patient samples obtained from the eyes without drops demonstrated a composition similar to that of the treated eye with Firmicutes (56.8%), Actinobacteriota (16.9%), and Verrucomicrobiota (16.8%) being the most abundant phyla and Akkermansia (16.8%), Corynebacterium (8.5%), Faecalibaterium (6.0%), Lachnospiraceae (5.1%), and Blautia (3.2%) making up the majority of the genus classifications (Fig. 3).
Figure 3.
Taxonomy bar plot of microbial composition in control and patient (“no drops” and “drops”) eye samples. Patient samples are divided into “Drops” for samples obtained from eyes receiving topical glaucoma medications and “No Drops” for samples obtained from contralateral eyes not receiving topical eyedrops. Only species with relative abundance >1.0% were represented in the figure and considered in pairwise comparisons. Relative abundance values for each individual sample were first calculated by dividing the number of read counts assigned to the amplicon sequence variants corresponding to individual microbes by the total read count of the sample. The relative abundance values represented in the figure are averages across all samples of the respective category (“Controls,” “No Drops,” “Drops”). (A) Composition of organisms based on relative abundance at the phyla level. Patient samples contained higher abundance of Firmicutes, Verrucomicrobiota, Proteobacteria, Patescibacteria, Bacteroidota, Fusobacteriota, and Cyanobacteria (P < 0.05, LDA > 2) compared to control samples. Control samples contained higher abundance of Actinobacteriota (P < 0.05, LDA > 2). (B) Composition of organisms based on relative abundance at the genus level. Patient samples had higher abundance of Akkermansia, Faecalibacterium, Lachnospiraceae genus, Lachnospiraceae NK4A136 group, Clostridia UCG-014, Gluconobacter, Ruminococcus torques group, Bifidobacterium, Clostridia vadin B860 group, Oscillospiraceae UCG-002, Peptoniphilus, and Rothia compared to control samples (P < 0.05, LDA > 2). Control samples had increased abundance in Corynebacterium and Propionibacteriaceae Cutibacterium (P < 0.05, LDA > 2). No differences in relative abundance were found between eyes treated with drops and untreated patient eye samples at both the phyla (A) and genus (B) level. LDA - linear discriminant analysis, statistical test used after performing centered log-ratio (CLR) transformation of raw read counts.
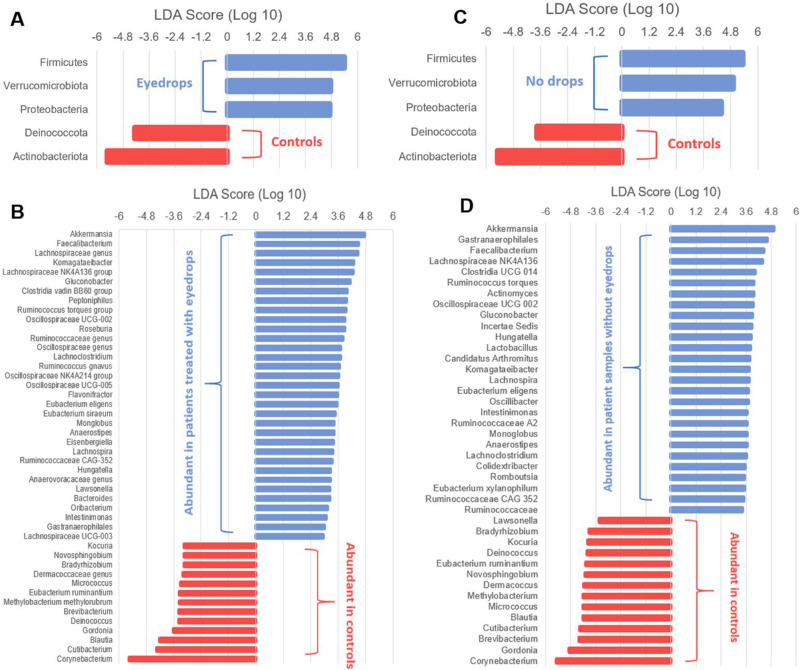
Linear discriminant analysis comparison of the normalized relative abundances of assigned taxonomic classifications identified differentially abundant organisms, with findings supplemented by comparisons of microbiome composition using ASVs performed by DeSeq2. We identified Firmicutes, Verrucomicrobiota, and Proteobacteria as the three phyla with increased abundance in patient eye samples irrespective of glaucomatous treatment, while control eye samples had increased abundance of Deinococcota and Actinobacteriota. At the genus level, the abundances of 33 and 27 bacterial taxa were increased in patient eyes with drops and eyes without drops compared to healthy controls, respectively (Fig. 4). Analysis with DeSeq2 identified 57 ASVs with increased abundance in patient eye samples with drops and 85 ASVs with increased abundance in patient eye samples without drops (Fig. 5) compared with healthy control samples. The differentially abundant taxa and ASVs demonstrated an increased presence of anaerobic, gram-negative organisms from the genus Akkermansia, Faecalibacterium, Lachnospiraceae, and Komagataeibacter on the ocular surface of patient eyes receiving topical medication compared to healthy controls that primarily consisted of gram-positive organisms of the genus Corynebacterium, Cutibacterium, Blautia, and Gordonia (Table 4, Supplemental Table S1). A similar pattern was observed for the ocular surface of the patient eye without drops with the increased abundance of gram-negative organisms from the genus Akkermansia, Gastranaerophilales, Faecalibacterium, and Lachnospiraceae NK4A136 group, as well as the presence of anaerobic, gram-positive organisms including Roseburia, Clostridia UCG-014, and Ruminococcus torques group (Supplemental Table S2). The mean number of sequence read counts was significantly lower for both patient eye samples with eyedrops (3,792.70) and without drops (3,898.95) compared to healthy controls (18,498.43, P < 0.0001).
Figure 4.
Linear discriminant analysis (LDA) effect size (LEfSe) plot of taxonomic biomarkers identified in the ocular surface microbiome of patients and controls. LDA scores are effect size estimates for a particular taxonomic marker in a specific group, with values interpreted as the relative magnitude of abundance compared to the other group. Specific bacterial sequence variants at the phyla (A, C) and genus (B, D) level were sorted based on patient eyes treated with eyedrops (A, B) and patient eyes without treatment (C, D) with healthy control samples used as the comparison group. Patient eyes treated with eyedrops (A) and untreated patient eyes (C) had greater abundance of Firmicutes, Verrucomicrobiota, and Proteobacteria and lower abundance of Deinococcota and Actinobacteriota compared to controls. The relatively abundant bacterial sequence variants at the genus level are labelled for eye samples with drops (B) and without drops (D) in blue and controls in red (B, D). All LDA scores and bacterial sequence variants presented are statistically significant, with P < 0.05 (Kruskal-Wallis and pairwise Wilcoxon test) and LDA score ≥ 2.0 the criteria for meeting significance.
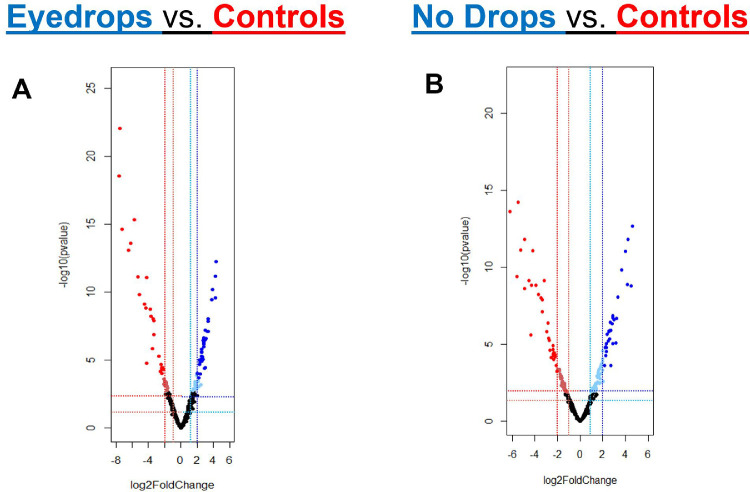
Figure 5.
Volcano Plots for ASVs. Sequence variants (each individual point) and their log2foldchange are plotted comparing (A) patient eye samples receiving eyedrop treatment compared to controls and (B) patient eye samples without eyedrop treatment compared to controls. The log2fold change values are outputs based on DeSEQ2 workflow. Sequence variants found to be relatively abundant with P < 0.05 and log2foldchange > 1.0 are denoted in light blue (relatively abundant in patients) and light red (relatively abundant in controls). Sequence variants found to be relatively abundant with P < 0.01 and log2foldchange > 2.0 are denoted in blue (relatively abundant in patients) and red (relatively abundant in controls). The sequence variants and taxonomic assignments associated with individual data points are noted in Supplemental Tables S1, S2.
Table 4.
Most Differentially Abundant Taxonomic Classifications Represented in Each Ocular Surface Sample Group
| Phyla | Genus | Species | Log2FC | P Value |
| Abundant Taxonomic Classifications in Patient Samples With Eyedrops | ||||
|---|---|---|---|---|
| Proteobacteria | Komagataeibacter | rhaeticus/xylinus | 4.363 | 1.77E-10 |
| Firmicutes | Eubacterium siraeum | .. | 4.238 | 1.87E-09 |
| Verrucomicrobiota | Akkermansia | muciniphila | 4.228 | 5.11E-08 |
| Firmicutes | Eubacterium eligens | .. | 3.861 | 1.46E-08 |
| Actinobacteriota | Bifidobacterium | bifidum | 3.765 | 6.45E-08 |
| Firmicutes | Ruminococcaceae CAG-352 | .. | 3.364 | 8.78E-06 |
| Firmicutes | Blautia | .. | 3.320 | 1.78E-06 |
| Firmicutes | Oscillospiraceae NK4A214 group | .. | 3.306 | 1.25E-06 |
| Firmicutes | Lachnospiraceae NK4A136 group | .. | 3.240 | 8.78E-06 |
| Firmicutes | Gaecalibacterium | prausnitzii | 3.042 | 2.47E-05 |
| Abundant taxonomic classifications in Patient Samples without Eyedrops | ||||
| Verrucomicrobiota | Akkermansia | muciniphila | 4.483 | 4.78E-08 |
| Firmicutes | Eubacterium siraeum group | .. | 4.209 | 9.76E-11 |
| Verrucomicrobiota | Akkermansia | muciniphila | 4.183 | 4.50E-08 |
| Proteobacteria | Gluconobacter | .. | 3.973 | 4.53E-10 |
| Actinobacteriota | Bifidobacterium | bifidum | 3.678 | 6.55E-09 |
| Firmicutes | Roseburia | intestinalis | 3.308 | 2.40E-07 |
| Firmicutes | Ruminococcus torques group | .. | 3.242 | 4.03E-06 |
| Actinobacteriota | Bifidobacterium | longum | 3.159 | 1.12E-04 |
| Firmicutes | Lachnospiraceae | .. | 3.000 | 4.78E-06 |
| Firmicutes | Blautia | .. | 2.877 | 3.15E-06 |
| Abundant taxonomic classifications in Control Subject Samples | ||||
| Actinobacteriota | Corynebacterium | aurimucosum/pseudogenitalium/tuberculostearicum | −5.142 | 3.10E-08 |
| Actinobacteriota | Corynebacterium | kroppenstedtii | −5.269 | 1.92E-09 |
| Actinobacteriota | Cutibacterium | acnes/avidum | −5.720 | 2.54E-13 |
| Actinobacteriota | Corynebacterium | bovis | −6.193 | 1.06E-11 |
| Actinobacteriota | Corynebacterium | kroppenstedtii | −6.468 | 3.09E-11 |
| Actinobacteriota | Corynebacterium | .. | −7.277 | 1.15E-12 |
| Actinobacteriota | Corynebacterium | .. | −7.556 | 7.82E-20 |
| Actinobacteriota | Corynebacterium | kroppenstedtii | −7.566 | 2.02E-16 |
| Actinobacteriota | Corynebacterium | macginleyi | −9.671 | 8.55E-23 |
| Actinobacteriota | Corynebacterium | bovis | −9.839 | 2.21E-21 |
The 10 taxonomic classifications with the greatest significance in differential relative abundance when performing pairwise comparisons are listed. Positive log fold change denotes increased abundance in patient samples and negative log fold change represents increased abundance in control subject samples. All P values are adjusted with Benjamini-Hochberg correction. The variants associated with each taxonomic classification is noted in Supplemental Tables 1, 2.
The observed shift from a predominant gram-positive composition of the healthy control subjects to the anaerobic and gram-negative composition observed in both the patients’ treated and contralateral untreated eyes were associated with reported ocular symptom scores (DEQ-5) and tear film measures. The ASVs that had significant positive associations with DEQ-5 score, suggesting increased symptoms of ocular irritation, were classified to organisms observed in greater abundance on the ocular surface of patients such as Clostridia and Lachnospiraceae. In addition, gram-negative organisms with increased abundance on patient eyes such as Oscillospiraceae, Lachnospiraceae (NK4A136 and Ruminococcus torques groups), Bifidobacterium, and Akkermansia were negatively associated with tear meniscus height. The gram-positive organisms that colonized the ocular surface of healthy controls such as Corynebacterium and Lawsonella were found to be negatively associated with DEQ-5 score and positively associated with tear meniscus height (Table 5; Supplemental Data S2–S5).
Table 5.
Taxonomic Classifications of ASVs Associated With DEQ-5 Scores and Tear Meniscus Height Measurements
| Drops vs. Controls | No Drops vs. Controls | |||
|---|---|---|---|---|
| DEQ5 score | Tear meniscus height | DEQ 5 Score | Tear meniscus height | |
| Positive association (increased abundance with higher measure) | Clostridia vadin BB60 group* | Corynebacterium aurimucosum | Lachnospiraceae NK4A136 group* | Monoglobus pectinilyticus * |
| Lachnospiraceae genus* | Knoellia aerolata/subterranea | Lawsonella sp. | Ruminococcus bicirculans * | |
| Lachnospiraceae NK4A136 group* | Neomicrococcus aestuarii | Bifidobacterium longum * | Knoellia aerolata/subterranea | |
| Fusobacterium sp. | Corynebacterium sp. | Clostridia vadin BB60 group* | Neomicrococcus aestuarii | |
| Agathobacter sp. * | Fusobacterium sp. | Lachnospiraceae Eubacterium xylanophilum group* | Corynebacterium sp. | |
| Negative association (decreased abundance with higher measure) | Lachnospiraceae Ruminococcus torques group* | Oscillospiraceae NK4A214 group* | Corynebacterium kroppenstedtii | Corynebacterium kroppenstedtii |
| Oscillospiraceae NK4A214 group* | Lachnospiraceae NK4A136 group* | Lawsonella sp. | Oscillospiraceae NK4A214 group* | |
| Corynebacterium kroppenstedtii | Agathobacter Eubacterium ramulus * | Corynebacterium sp. | Ruminococcus callidus * | |
| Roseburia inulinivorans | Bifidobacterium sp. * | Kocuria sp. | Akkermansia muciniphila * | |
| Lachnospira sp. * | Roseburia hominis | Incertae-sedis Clostridium sp.* | Lachnospiraceae Ruminococcus torques group* | |
The top five ASVs with a positive association (defined as a positive log-fold change with increasing continuous variable) and negative association (defined as a negative log-fold change with increasing continuous variable) are listed for two pairwise comparisons. “Drops vs. Controls” represents patient eye samples treated with topical eyedrops compared to healthy controls and “No Drops vs. Controls” represents patient eye samples not treated with eyedrops compared to healthy controls. The treatment group was controlled for when testing for association with the continuous variables: DEQ-5 score and tear meniscus height. All taxonomic classifications noted are significantly associated with DEQ5 score and tear meniscus height measures with Benjamini-Hochberg corrected P < 0.01. The full dataset for the table can be found in Supplemental Data 1 and Supplemental Data 2.
ASVs are significantly more abundant in patient eye samples compared to controls.
Metagenome Inference Analysis Suggests Ocular Surface Microbiota of Patients Using Glaucoma Drops Are More Capable of Lipopolysaccharide Synthesis Whereas Microbiota of Healthy Controls Have Capacity for Reducing Inflammation
PiCRUST2 was used for the prediction of metagenome functions generated from the ASV table and the raw sequencing reads, identifying 793 differentially abundant Enzyme Classification (EC) libraries, 2208 KEGG orthologs, and 137 MetaCyc pathways between patient eye samples treated with drops compared to healthy controls. A similar amount of variation in function was seen when comparing the predicted metagenomes for patient eye samples without drops and healthy controls with 747 EC libraries, 2059 KEGG orthologs, and 128 MetaCyc pathways identified as differentially abundant (Supplemental Data S6–S9). There were no differences in the relative composition of EC libraries, KEGG orthologs, and MetaCyc pathways between the treated eye and untreated eye for patient samples.
The shared metagenome pathways for the patient eye samples, irrespective of treatment status, were predominantly related to lipopolysaccharide (LPS) synthesis and included GDP-mannose-derived O-antigen biosynthesis, lipid IVa biosynthesis, 3-deoxy-D-manno-octulosonate biosynthesis I, preQ0 biosynthesis, and D-manno-heptose biosynthesis (Supplemental Table S3). The abundance of LPS synthesis pathways were found to be associated with sequence variants classified to Bacteroides, Bradyrhizobium, Christensenellaceae R-7 group, Gluconobacter, Komagataeibacter, Akkermansia, and Lachnospiraceae groups, with the latter two mentioned previously for associations with DEQ-5 score and tear meniscus height. In addition, pathways for anaerobic unsaturated and saturated fatty acid synthesis (gondoate biosynthesis, cis-vaccenate biosynthesis) and hydrogen sulfide and sulfate metabolism (sulfate reduction I, sulfate assimilation and cysteine biosynthesis, L-methionine biosynthesis) were significantly increased in abundance for patient eye samples, with contributions from Akkermansia, Lachnospiraceae groups, Ruminococcus, and Oscillospiraceae groups. For the healthy control eye samples, the abundant pathway functions were related to carbohydrate synthesis, glycolysis, and oxidative phosphorylation (glyoxylate cycle, glyoxylate bypass and TCA, glycolysis, pyruvate dehydrogenase, aerobic respiration I cytochrome c) with other pathways involved in the biosynthesis of mycothiol, biotin, and heme. These factors detoxify alkylating agents, reactive oxygen and nitrogen species,34,35 and modulate inflammation36 and were attributed to Corynebacterium, Cutibacterium, Methylobacterium, Rothia, Lawsonella, Brevibacterium, and Kocuria.
Discussion
This study demonstrates that the ocular surface microbiome of patients taking unilateral preserved glaucoma drops is characterized by a highly diverse array of gram-negative bacteria that is significantly different from the predominantly gram-positive microbes detected on healthy control eyes. An unexpected finding was observing similar changes to the ocular surface microbiota in both the asymmetric/unilateral glaucoma patients’ treated and untreated eyes. Because of the design of the study, it is impossible to determine whether the microbial changes were related to the drop used to treat glaucoma or to the diagnosis of glaucoma itself. The patients studied all had highly asymmetric or truly unilateral (e.g., traumatic, angle closure) glaucoma, and yet both eyes had similar microbiome findings, which were distinct from drop-naïve, control subjects without any diagnosis of glaucoma.
Several studies implicate extra-ocular microbiome alterations in open angle glaucoma pathogenesis. Although higher incidences of gastric infections with the gram-negative Helicobacter pylori have been demonstrated in patients with open-angle glaucoma versus non-glaucomatous controls,37–41 other studies have failed to confirm this association.42 Case-control studies43 have found that patients with open angle glaucoma have greater oral bacterial loads than controls including more Streptococcus sp. and fewer natural teeth.44 Patients with new-onset open-angle glaucoma have higher incidences of tooth loss in the two years leading up to their glaucoma diagnosis.45 Astafurov and colleagues43 postulate that pre-existing oral microbiome dysbiosis contributes to the pathogenesis of glaucoma by lipopolysaccharide-driven upregulation of toll-like receptor 4 and the complement system in the retina. None of these studies have investigated whether the glaucoma therapies used by these patients, including preserved drops, contributed to the observed orogastric microbial alterations. To our knowledge, no studies have been performed comparing oral or ocular surface microbiomes in therapy-naïve glaucoma patients to age- and sex-matched controls.
Another potential explanation for our findings is that exposure to preserved eyedrops in one eye may be sufficient to drive similar changes in the contralateral eye. Prior studies comparing left and right ocular surface microbiota have found no differences in composition or diversity.46,47 The ocular surface microbiome of two paired eyes may act as a singular microbial ecosystem rather than two independent systems. It is possible that similarities between the microbiomes of left and right eyes may be due to mechanical transfer of organisms by eye-rubbing or via the lacrimal and nasal mucosa linking the left and right conjunctiva. Further work is needed to explore the possibility of binocular microbiome shifts associated with uniocular perturbations.
Honda and colleagues48 similarly found that the conjunctivae of patients receiving topical glaucoma eyedrops had increased abundance of gram-negative organisms compared to healthy controls, although the study was restricted to culturable microbes. Another study49 using culture-based methods found that conjunctival microbial isolates from glaucoma patients using 0.005% latanoprost preserved with 0.02% BAK contained more antibiotic-resistant Staphylococcus epidermidis than conjunctival isolates from patients using 0.004% travoprost preserved with a zinc-based ion buffer system. Recently, Lim and colleagues50 described the eyelid margin and buccal microbiomes obtained via 16S rRNA sequencing of 30 open angle glaucoma (OAG) patients treated with preserved prostaglandin analogs compared with 32 eyedrop-naïve, recently diagnosed OAG patients. The authors found that the eyelid microbiome of patients receiving preserved prostaglandin was enriched for Azomonas, Pseudomonas, and Granulicatella with relative depletion of Delftia and Rothia. Both alpha- and beta-diversity were increased in eyelid margin and buccal mucosa microbiotas of OAG patients receiving topical prostaglandin therapy versus drop-naïve patients. Although the study by Lim et al.50 supports our hypothesis that preserved drops rather than the diagnosis of glaucoma affects the ocular surface microbiome, the microbial compositional differences between groups in Lim's study were less pronounced than in ours.
BAK is the most frequently found preservative used in glaucoma eye drops and artificial tears.9,51 At low concentrations, BAK primarily inhibits gram positive organisms52 with increased activity against gram negative organisms at higher concentrations or in the presence of EDTA.6 In our study, patient eyes were associated with a decreased presence of gram-positive colonizers of the ocular surface such as Corynebacterium and Cutibacterium, and, instead, were associated with predominantly gram-negative organisms consisting of Akkermansia, Faecalibacterium, and Lachnospiraceae groups that are often found in the human gut microbiome. Our patient population was treated with a variety of eyedrop formulations containing different active ingredients yet exhibited similar ocular microbiome changes. This suggests that if eyedrops are acting on the ocular surface microbiome, the active medication ingredient (e.g., prostaglandin analog, beta blocker, alpha agonist) may not be responsible for inducing the changes but rather an ingredient common to all the drops, such as the preservative.
The presence of BAK persists on the ocular surface with a half-life of 20 hours in corneal epithelial tissues and 11 hours in deeper conjunctival layers,53 with detectable concentrations up to one week after instillation.54 These pharmacokinetics are consistent with a hypothesis that chronic daily exposure to low concentrations of BAK found in preserved ophthalmic drops can generate persistent, rather than transient, changes to the ocular surface microbiome. Disruption of the tear film by BAK9 may also create a local hypoxic ecological niche that preferentially selects for gram-negative anaerobes, as the aqueous layer of the tear film is essential for the transfer of oxygen and nutrients across the ocular surface.55
The use of BAK in eyedrop formulations has been attributed to symptoms of ocular surface irritation that can affect compliance with long-term therapy.5–8,56,57 In our study, the patient eye samples exposed to preserved eyedrops had significantly lower tear meniscus height and first tear break-up time compared to patient eyes without drops and healthy controls, suggesting an ocular surface that is more susceptible to desiccating stress. Patients also reported increased ocular irritative symptoms in the treated versus untreated eye.
In our study, ocular samples obtained from healthy control subjects had a higher abundance of Corynebacterium (71.7%) compared to healthy participants reported in much of the prior literature (range 8.0%–28.2%).16–18 In these studies, a large proportion of the healthy cohort came from volunteers who were visitors to clinical ophthalmology clinics and were significantly younger than our cohort, with majority being under 60 years old. The microbiota composition of our older healthy cohort more closely matched volunteers randomly selected from the community in studies by Kang et al.,58 where they found four healthy controls with high abundances of Corynebacterium (53.9%–78.5%), and Suzuki et al.,59 with healthy participants greater than the age of 60 having a microbiome predominantly consisting of Corynebacterium. Although compositional differences were observed between studies based on recruitment methods and cohort age, the ocular surface microbiomes of healthy participants in these studies were consistently colonized by gram-positive organisms.
Disruption of gram-positive communities may create an ocular surface at greater risk for infection and inflammation. A study performed on a mouse model of ocular surface disease described Corynebacterium mastiditis as a stable colonizer of the ocular surface that is capable of inducing a T-cell mediated response to prevent pathogenic colonization of Candida albicans and Pseudomonas aeruginosa.60 Previous studies characterizing the ocular microbiome in dry eye disease patients demonstrated decreased abundance of gram-positive organisms including coagulase-negative Staphylococcus, Corynebacterium, and Streptococcus,61 along with an increased abundance of gram-negative organisms from the genus Pseudomonas, Bacteroidetes, Bradyrhizobium, and Bifidobacterium.62 It is unclear whether dry eye patients in these studies were routinely using preserved artificial tear drops, which might have altered the ocular surface microbiome.
Research studies that characterize the ocular surface microbiome in the setting of contact lens use provide additional perspective as contact lens wash solutions also contain anti-microbial chemicals. Peroxide-based contact lens solutions have been shown to significantly decrease the abundance of gram-positive microbes Corynebacterium and Streptococcus.63 Contact lens wearers exhibit significantly higher abundance of gram-negative organisms including Pseudomonas, Acinetobacter, Methylobacterium, and Lactobacillus with decreased abundance of Haemophilus, Streptococcus, Staphylococcus, and Corynebacterium.64 These microbial compositional differences match closely with findings from our study. The increased colonization of gram-negative organisms has been previously associated with increased number of cornea infiltrative events,65 suggesting that the replacement of protective gram-positive strains with gram-negative organisms may predispose patients to ocular infections.
Although both eyes were colonized by similar microbes in our asymmetric glaucoma patient group, the eyes treated with glaucoma drops exhibited greater dry eye symptoms compared to the contralateral untreated eye, suggesting that the irritative effect may be caused by the eyedrop itself, and is consistent with the literature.66 However, our study also hints at the possibility that microbiome changes could potentiate these effects. Metagenome inference analysis (PiCRUST2) suggests that the abundant gram-negative organisms associated with patient samples are capable of synthesizing LPS, a component of the cytoplasmic membrane that functions as an endotoxin binding to toll-like receptors of immune cells. On the ocular surface, LPS has been shown to interact with toll-like receptor 4 resulting in the downstream production of cytokines and microglial activation67 with direct ocular exposure to LPS linked to increased inflammatory transcripts IL-1β, TNF-α, and CXCL10 in the cornea and conjunctiva.68
A final and scientifically less interesting explanation for similar microbiomes in treated and untreated patient eyes but differences between patients and controls is that there was no true effect of the preserved eyedrops on the ocular surface microbiome but some nontrivial difference between the patients and controls in terms of their underlying characteristics or in the way their samples were collected or processed. To reduce confounding biases, we utilized age-matched community controls from the same geographic location. The same sampling and processing techniques were employed by the same study personnel throughout. With paucibacterial sites such as the ocular surface, 16S rRNA microbiome sequencing is susceptible to significant artefact and contamination. Doan and colleagues18 demonstrated that false identification of genera due to PCR amplification errors can quickly outnumber true genera when sequencing very low DNA burden specimens. “Air swab” negative controls were used in our study to reduce potential batch effect and influence of systematic DNA contaminants which may arise during sample collection and processing. The use of mock bacterial communities as positive controls has been proposed at the recent National Eye Institute Anterior Segment Initiative Symposium on Ocular Surface Microbiome Best-Practices for Low Biomass Research and would have allowed us to be more confident in our 16S results.69 Last, there is the possibility that our controls are somehow different from our patients in a significant but obscure way for which we did not control.
Our study has several limitations. We accounted for the limited cohort size by obtaining multiple samples from each subject and adjusting for repeat measures. However, the study design remains susceptible to sampling bias as there was a wide range of feature counts from samples obtained from the same subject, which may not provide an accurate or complete representation of the study population. Variable feature counts also suggest differential abundance in sample biomass that can significantly affect alpha and beta-diversity analysis.70 A proposed methodology to standardize the concentration of samples has been documented by Doan and colleagues.18 A limitation to our methodology was not performing quantitative polymerase chain reaction (qPCR) to control for sample concentration. In addition, qPCR has the added benefit of confirming the presence of detected microbes using species-specific primers outside of the 16S region which we propose as a helpful addition in future studies. There is currently a lack of standardization of contaminant filtering for low burden microbial niches. In addition to the filtering methods we used, other pipelines have been proposed and could be used as alternative methodology.71
Hypothesis-generating case-control studies such as ours are useful in testing for associations that can support novel theoretical mechanisms of disease but are limited in their ability to prove causation. As stated before, our study was not designed to conclusively differentiate between effects on the ocular surface microbiome from preserved eye drops versus from the underlying glaucoma. Non-living bacteria having arrived on the ocular surface from the environment cannot be discriminated from alive colonizers using 16S rRNA sequencing. Shotgun DNA sequencing has an advantage over 16S as it allows for species and strain level resolution rather than genera, as well as the identification of DNA viruses and fungi. Metagenome inference analysis (PiCRUST2) is imperfect at predicting function of 16S microbiomes as it is only a best guess of functional capacity based on the pooled known functions of sequenced genera. RNA-seq can be used to identify actual microbial mRNA transcripts and, when coupled with shotgun sequencing, allows for not only identification of which specific microbial species and strains are present but also the specific proteins they are actively producing.
In summary, the ocular surface microbiome of patients taking unilateral preserved glaucoma drops is characterized by a highly diverse array of gram-negative bacteria that is significantly different from the predominantly gram-positive microbes found on healthy control eyes. These compositional differences were associated with decreased tear film measures and distinct inferred protein synthesis pathways, suggesting a potential link between microbial alterations and ocular surface disease. Future prospective, randomized, and masked studies using shotgun metagenomics, RNA-seq, and other functional assays will be needed to understand the interplay between topical medications, the ocular surface microbiome, and inflammation in the setting of glaucoma.
Supplementary Material
Acknowledgments
Special thanks to Kathryn McCauley, PhD, Bing Zhang, MD, and Preston Tasoff for their reviews of the manuscript prior to submission.
Supported, in part, by the UCSF Vision Shared Resource Core Grant (NIH/NEI P30 EY002162) and departmental support from Research to Prevent Blindness to both the UCSF and Columbia University departments of ophthalmology.
Disclosure: C.-C.J. Chang, None; K. Somohano, None; C. Zemsky, None; A.-C. Uhlemann, None; J. Liebmann, None; G.A. Cioffi, None; L.A. Al-Aswad, None; S.V. Lynch, Siolta Therapeutics, Inc. (E, I, C); B.J. Winn, None
References
- 1. Liu SA, Zhao ZN, Sun NN, Han Y, Chen J, Fan ZG.. Transitions of the understanding and definition of primary glaucoma. Chin Med J. 2018; 131: 2852–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halpern DL, Grosskreutz CL.. Glaucomatous optic neuropathy: mechanisms of disease. Ophthalmol Clin North Am. 2002; 15(1): 61–68. [DOI] [PubMed] [Google Scholar]
- 3. Dunn N, Mullee M, Perry H, Holmes C.. Association between dementia and infectious disease: evidence from a case-control study. Alzheimer Dis Assoc Dis. 2005; 19: 91–94. [DOI] [PubMed] [Google Scholar]
- 4. Tanito M, Kaidzu S, Takai Y, Ohira A.. Association between systemic oxidative stress and visual field damage in open-angle glaucoma. Sci Rep. 2016; 6: 25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaenen N, Baudouin C, Pouliquen P, Manni G, Figueiredo A, Zeyen T.. Ocular symptoms and signs with preserved and preservative-free glaucoma medications. Eur J Ophthalmol. 2007; 17: 341–349. [DOI] [PubMed] [Google Scholar]
- 6. Baudouin C, Labbe A, Liang H, Pauly A, Brignole-Baudouin F.. Preservatives in eyedrops: the good, the bad and the ugly. Prog Retin Eye Res . 2010; 29: 312–334. [DOI] [PubMed] [Google Scholar]
- 7. Fechtner RD, Godfrey DG, Budenz D, Stewart JA, Stewart WC, Jasek MC.. Prevalence of ocular surface complaints in patients with glaucoma using topical intraocular pressure-lowering medications. Cornea. 2010; 29: 618–621. [DOI] [PubMed] [Google Scholar]
- 8. Henry JC, Peace JH, Stewart JA, Stewart WC.. Efficacy, safety, and improved tolerability of travoprost BAK-free ophthalmic solution compared with prior prostaglandin therapy. Clin Ophthalmol. 2008; 2: 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yalvac IS, Gedikoglu G, Karagoz Y, et al.. Effects of antiglaucoma drugs on ocular surface. Acta Ophthalmologica Scandinavica. 1995; 73(3): 246–248. [DOI] [PubMed] [Google Scholar]
- 10. Kuppens EV, de Jong CA, Stolwijk TR, de Keizer RJ, van Best JA.. Effect of timolol with and without preservative on the basal tear turnover in glaucoma. Br J Ophthalmol . 1995; 79: 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Jong C, Stolwijk T, Kuppens E, de Keizer R, van Best J.. Topical timolol with and without benzalkonium chloride: epithelial permeability and autofluorescence of the cornea in glaucoma. Graefes Arch Clin Exp Ophthalmol . 1994; 232: 221–224. [DOI] [PubMed] [Google Scholar]
- 12. Herreras JM, Carlos Pastor J, Calonge M, Asensio VM. Ocular surface alteration after long-term treatment with an antiglaucomatous drug. Ophthalmology. 1992; 99: 1082–1088. [DOI] [PubMed] [Google Scholar]
- 13. Sherwood MB, Grierson I, Millar L, Hitchings RA.. Long-term morphologic effects of antiglaucoma drugs on the conjunctiva and Tenon's capsule in glaucomatous patients. Ophthalmology. 1989; 96: 327–335. [DOI] [PubMed] [Google Scholar]
- 14. Mietz H, Niesen U, Krieglstein GK.. The effect of preservatives and antiglaucomatous medication on the histopathology of the conjunctiva. Graefes Arch Clin Exp Ophthalmol . 1994; 232: 561–565. [DOI] [PubMed] [Google Scholar]
- 15. Ammar DA, Noecker RJ, Kahook MY.. Effects of benzalkonium chloride-preserved, polyquad-preserved, and sofZia-preserved topical glaucoma medications on human ocular epithelial cells. Adv Ther. 2010; 27: 837–845. [DOI] [PubMed] [Google Scholar]
- 16. Dong Q, Brulc JM, Iovieno A, et al.. Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci . 2011; 52: 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang Y, Yang B, Li W.. Defining the normal core microbiome of conjunctival microbial communities. Clin Microbiol Infect. 2016; 22: 643.e7–643.e12. [DOI] [PubMed] [Google Scholar]
- 18. Doan T, Akileswaran L, Andersen D, et al.. Paucibacterial microbiome and resident DNA virome of the healthy conjunctiva. Invest Ophthalmol Vis Sci . 2016; 57: 5116–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kugadas A, Gadjeva M.. Impact of microbiome on ocular health. Ocular Surface. 2016; 14: 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrillo F, Pignataro D, Lavano MA, et al.. Current evidence on the ocular surface microbiota and related diseases. Microorganisms. 2020; 8: 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klindworth A, Pruesse E, Schweer T, et al.. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013; 41(1): e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park JY, Kim BG, Kim JS, Hwang JH.. Matrix metalloproteinase 9 point-of-care immunoassay result predicts response to topical cyclosporine treatment in dry eye disease. Transl Vis Sci Technol . 2018; 7(5): 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chalmers RL, Begley CG, Caffery B.. Validation of the 5-item Dry Eye Questionnaire (DEQ-5): discrimination across self-assessed severity and aqueous tear deficient dry eye diagnoses. Cont Lens Anterior Eye. 2010; 33(2): 55–60. [DOI] [PubMed] [Google Scholar]
- 24. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP.. DADA2:High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016; 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolyen E, Rideout JR, Dillon MR, et al.. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019; 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quast C, Pruesse E, Yilmaz P, et al.. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41(D1): D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R.. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010; 5: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R.. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013; 2(1): 2047–217X–2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vazquez-Baeza Y, Gonzalez A, Smarr L, et al.. Bringing the dynamic microbiome to life with animations. Cell Host Microbe. 2017; 21: 7–10. [DOI] [PubMed] [Google Scholar]
- 30. Segata N, Izard J, Waldron L, Gevers D, et al.. Metagenomic biomarker discovery and explanation. Genome Biology. 2011; 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douglas GM, Maffei VJ, Zaneveld JR, et al.. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020; 38: 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fernandes AD, Reid JN, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB.. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pedre B, Van Molle I, Villadangos AF, et al.. The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Mol Microbiol . 2015; 96: 1176–1191. [DOI] [PubMed] [Google Scholar]
- 35. Shinjyo N, Kita K.. Relationship between reactive oxygen species and heme metabolism during the differentiation of Neuro2a cells. Biochem Biophys Res Commun . 2007; 358: 130–135. [DOI] [PubMed] [Google Scholar]
- 36. Agrawal S, Agrawal A, Said HM.. Biotin deficiency enhances the inflammatory response of human dendritic cells. Am J Physiol Cell Physiol . 2016; 311(3): C386–C391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kountouras J, Mylopoulos N, Boura P, et al.. Relationship between Helicobacter pylori infection and glaucoma. Ophthalmology. 2001; 108(3): 599–604. [DOI] [PubMed] [Google Scholar]
- 38. Kountouras J, Zavos C, Chatzopoulos D.. Helicobacter pylori and glaucoma. Ophthalmology. 2003; 110: 2433–2434. [DOI] [PubMed] [Google Scholar]
- 39. Kountouras J, Mylopoulos N, Konstas AG, Zavos C, Chatzopoulos D, Boukla A.. Increased levels of Helicobacter pylori IgG antibodies in aqueous humor of patients with primary open-angle and exfoliation glaucoma. Graefes Arch Clin Exp Ophthalmol . 2003; 24: 884–890. [DOI] [PubMed] [Google Scholar]
- 40. Deshpande N, Lalitha P, Krishna das SR, et al.. Helicobacter pylori IgG antibodies in aqueous humor and serum of subjects with primary open angle and pseudo-exfoliation glaucoma in a South Indian population. J Glaucoma. 2008; 17: 605–610. [DOI] [PubMed] [Google Scholar]
- 41. Zeng J, Liu H, Liu X, Ding C.. The relationship between Helicobacter pylori infection and open-angle glaucoma: a meta-analysis. Invest Ophthalmol Vis Sci . 2015; 56: 5238–5245. [DOI] [PubMed] [Google Scholar]
- 42. Zullo A, Ridola L, Hassan C, Bruzzese V, Papini F, Vaira D.. Glaucoma and Helicobacter pylori: eyes wide shut? Dig Liver Dis . 2012; 44: 627–628. [DOI] [PubMed] [Google Scholar]
- 43. Astafurov K, Elhawy E, Ren L, et al.. Oral microbiome link to neurodegeneration in glaucoma. PLoS One. 2014; 9(9): e104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polla D, Astafurov K, Hawy E, Hyman L, Hou W, Danias J.. A pilot study to evaluate the oral microbiome and dental health in primary open-angle glaucoma. J Glaucoma. 2017; 26: 320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pasquale LR, Hyman L, Wiggs JL, et al.. Prospective study of oral health and risk of primary open-angle glaucoma in men. Ophthalmology. 2016; 123: 2318–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cavuoto KM, Mendez R, Miller D, Galor A, Banerjee S.. Effect of clinical parameters on the ocular surface microbiome in children and adults. Clin Ophthalmol . 2018; 12: 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okonkwo A, Rimmer V, Walkden A, et al.. Next-generation sequencing of the ocular surface microbiome: in health, contact lens wear, diabetes, trachoma, and dry eye. Eye Contact Lens. 2020; 46: 254–261. [DOI] [PubMed] [Google Scholar]
- 48. Honda R, Toshida H, Suto C, et al.. Effect of long-term treatment with eyedrops for glaucoma on conjunctival bacterial flora. Infect Drug Resist . 2011; 4: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ohtani S, Shimizu K, Neijima R, et al.. Conjunctival bacteria flora of glaucoma patients during long-term administration of prostaglandin analog drops. Invest Ophthalmol Vis Sci . 2017; 58: 3991–3996. [DOI] [PubMed] [Google Scholar]
- 50. Lim SH, Shin JH, Lee JW, Lee Y, Seo JH.. Differences in the eyelid and buccal microbiome of glaucoma patients receiving long-term administration of prostaglandin analog drops. Graefes Arch Clin Exp Ophthalmol . 2021; 259: 3055–3065. [DOI] [PubMed] [Google Scholar]
- 51. Kallings L, Rigertz O, Silverstolpe I.. Microbial contamination of medical preparations. Acta Pharm Suec . 1966; 3: 219–228. [PubMed] [Google Scholar]
- 52. McDonnell G, Russell AD.. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev . 1999; 12: 147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Champeau EJ, Edelhauser HF.. The effect of ophthalmic preservatives on the ocular surface: conjunctival and corneal uptake and distribution of benzalkonium chloride and chlorhexidine digluconate. In: Holly, F.J.(Ed.). The Preocular Tear Film in Health, Disease and Contact Lens Wear . Lubbock, TX: Dry Eye Institute, Inc.; 1986: 292–302. [Google Scholar]
- 54. Rolando M, Crider JY, Kahook MY.. Ophthalmic preservatives: focus on polyquaternium-1. Expert Opin Drug Deliv . 2011; 8: 1425–1438. [DOI] [PubMed] [Google Scholar]
- 55. Dartt DA, Willcox MDP.. Complexity of the tear film: Importance in homeostasis and dysfunction during disease. Exp Eye Res . 2013; 117: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Epstein SP, Ahdoot M, Marcus E, Asbell PA.. Comparative toxicity of preservatives on immortalized corneal and conjunctival epithelial cells. J Ocul Pharmacol Ther . 2009; 25: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pisella PJ, Pouliquen P, Baudouin C.. Prevalence of ocular symptoms and signs with preserved and preservative free glaucoma medication. Br J Ophthalmol . 2002; 86: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang Y, Zhang H, Hu M, et al.. Alterations in the ocular surface microbiome in traumatic corneal ulcer patients. Invest Ophthalmol Vis Sci. 2020; 61(6): 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki T, Sutani T, Nakai H, Shirahige K, Kinoshita S.. The microbiome of the meibum and ocular surface in healthy subjects. Invest Ophthalmol Vis Sci . 2020; 61(2): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. St Leger AJ, Desai JV, Drummond RA, et al.. An ocular commensal protects against corneal infection by driving an interleukin-17 response from mucosal γδ T cells. Immunity. 2017; 47(1): 148–158.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watters GA, Turnbull PR, Swift S, Petty A, Craig JP.. Ocular surface microbiome in meibomian gland dysfunction. Clin Exp Ophthalmol . 2017; 45: 105–111. [DOI] [PubMed] [Google Scholar]
- 62. Willis KA, Postnikoff CK, Freeman A, et al.. The closed eye harbours a unique microbiome in dry eye disease. Sci Rep. 2020; 10: 12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Retuerto MA, Szczotka-Flynn LO, Mukherjee PK, et al.. Diversity of ocular surface bacterial microbiome adherent to worn contact lenses and bacterial communities associated with care solution use. Eye Contact Lens. 2019; 45: 331–339. [DOI] [PubMed] [Google Scholar]
- 64. Shin H, Price K, Albert L, Dodick J, Park L, Dominguez-Bello MG.. Changes in the eye microbiota associated with contact lens wearing. mBio. 2016; 7(2): e00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sankaridurg PR, Sharma S, Willcox M, et al.. Bacterial colonization of disposable soft contact lenses is greater during corneal infiltrative events than during asymptomatic extended lens wear. J Clin Microbiol . 2000; 38: 4420–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Perez-Bartolome F, Martinez-de-la-Casa JM, Arriola-Villalobos P, Fernandez-Perez C, Polo V, Garcia-Feijoo J.. Ocular surface disease in patients under topical treatment for glaucoma. Eur J Ophthalmol . 2017; 27: 694–704. [DOI] [PubMed] [Google Scholar]
- 67. Rowan S, Taylor A.. The role of microbiota in retinal disease. Adv Exp Med Biol. 2018; 1074: 429–435. [DOI] [PubMed] [Google Scholar]
- 68. Wang C, Schaefer L, Bian F, et al.. Dysbiosis modulates ocular surface inflammatory response to liposaccharide. Invest Ophthalmol Vis Sci . 2019; 60: 4224–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Neuhold LA, Redford M, Van Gelder RN, Nelson K. National Eye Institute Anterior Segment Initiative Symposium: Ocular Surface Microbiome - Best Practices for Low Biomass Research. Available at: https://www.nei.nih.gov/sites/default/files/2021-10/MRDHNEI%20Microbiome%20Report%20FINAL.pdf. Retrieved May 19, 2022.
- 70. Villette R, Autaa G, Hind S, Holm JB, Moreno-Sabater A, Larsen M.. Refinement of 16S rRNA gene analysis for low biomass biospecimens. Sci Rep. 2021; 11: 10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M.. Temporal stability and composition of the ocular surface microbiome. Sci Rep. 2017;9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.