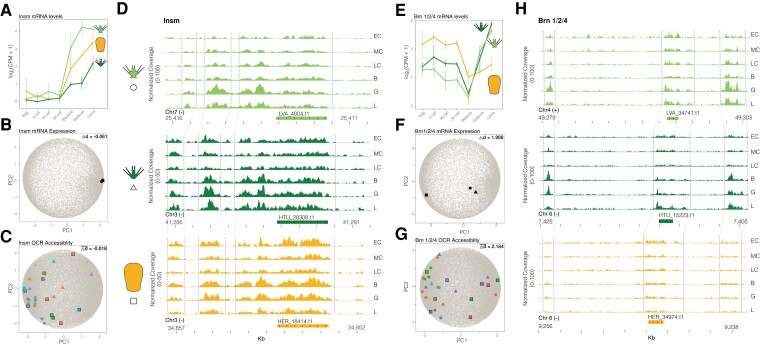
Fig. 8.
Quantifying developmental expression and accessibility changes in principal component space predicts divergent patterns of gene regulation. (A-D) An example of conserved developmental gene expression and chromatin accessibility for the gene insm. (A) Developmental mRNA expression profile of insm for each sea urchin species and (B) their coordinates in PCA space. (C) Coordinates in PCA space of developmental accessibility of orthologous OCRs near insm and (D) normalized chromatin accessibility coverage of these same OCRs through embryogenesis for each species. (E-H) An example of divergent developmental gene expression and chromatin accessibility associated with life history for the gene brn 1/2/4. (E) Developmental mRNA expression profile of brn 1/2/4 for each sea urchin species and (F) their coordinates in PCA space. (G) Coordinates in PCA space of developmental accessibility of orthologous OCRs near brn 1/2/4 and (H) normalized chromatin accessibility coverage of these same OCRs through embryogenesis for each species. See Supplementary Figure 6 for a summary figure of PCA distances for all genes and OCRs. OCR: open chromatin region; PCA: principal component analysis.