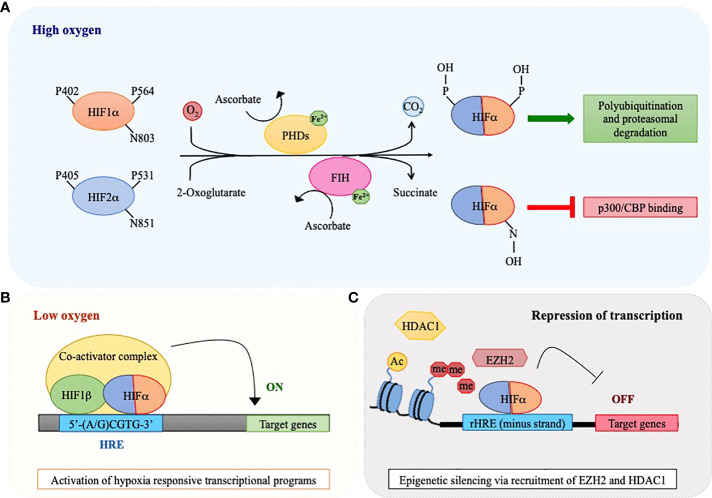
Figure 1.
Oxygen-sensing regulatory pathway of HIF1α and HIF2α. A schematic view of HIF1α and HIF2α regulation via the action of oxygen sensors prolyl hydroxylases (PHDs) and factor inhibiting HIF (FIH). (A) PHD and FIH are HIFα hydroxylases that require iron (Fe2+) and ascorbate as co-factors and utilize 2-oxoglutarate and molecular oxygen as co-substrates of their enzymatic reaction, with the release of CO2 and succinate as waste products. In the presence of oxygen, these enzymes catalyze hydroxylation of HIF1α and HIF2α at proline (P) and asparagine (N) residues respectively, thus provoking a dual effect: PHDs cause HIFα polyubiquitination and degradation by the proteasome, while FIH inhibits binding of co-activators like CREB-binding protein (CBP) and p300 to the HIF transcriptional complex. (B) In conditions of oxygen scarcity, the activities of PHDs and FIH are inhibited and HIFα subunits become stabilized, dimerize with HIF1β and bind hypoxia responsive elements (HREs) within the regulatory regions of specific target genes to activate their transcription. (C) HIFα factors may also recognize HREs in the minus DNA strand (reverse HRE, rHRE) thus allowing recruitment of histone methyltransferases or deacetylases (EZH2 and HDAC1) and provoking epigenetic silencing and transcriptional repression.