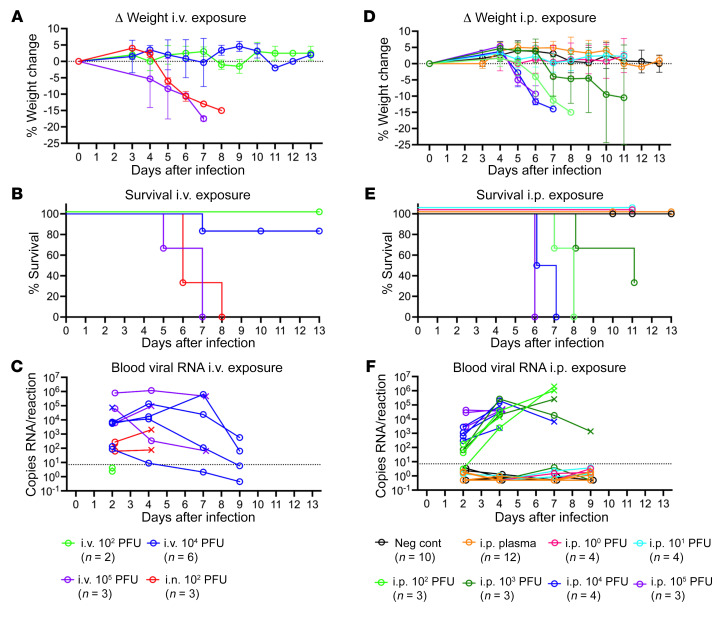
Figure 6. No infection observed with exposure to SARS-CoV2 RNA–positive human plasma.
(A–C) K18-hACE2–Tg IFNAR-KO mice were infected with 1.1 × 102 PFU, 1.1 × 104 PFU, or 1.1 × 105 PFU intravenously, or 1.1 × 102 PFU intranasally with the B1.1.7 variant of SARS-CoV-2. (D–F) K18-hACE2–Tg IFNAR-KO mice were infected with 1.1 × 100 PFU, 1.1 × 101 PFU, 1.1 × 102 PFU, 1.1 × 103 PFU, 1.1 × 104 PFU, or 1.1 × 105 PFU intraperitoneally with the B1.1.7 variant of SARS-CoV-2 or given 500 μL SARS-CoV2 RNA–positive human plasma intraperitoneally (6 samples into 2 mice each were tested). Unexposed IFNAR-KO littermates that do not carry the K18-hACE gene (noncarriers) were used as negative controls. (A and D) Weights were measured daily and the percentage weight change was calculated for each mouse over time, with mean change in weights and standard deviation plotted for each group. (B and E) Percentage survival over time by group. (C and F) At indicated time points, 20 μL of EDTA whole blood was collected, RNA was isolated, and SARS-CoV2 RNA levels were measured by qRT-PCR. Values are plotted for each mouse. × indicates nonsurviving mouse. Dashed line indicates maximum value detected among 63 negative blood sample controls plus 0.5 and was used as a cutoff for positive signal. When no viral RNA was detected, a value of 0.5 was assigned.