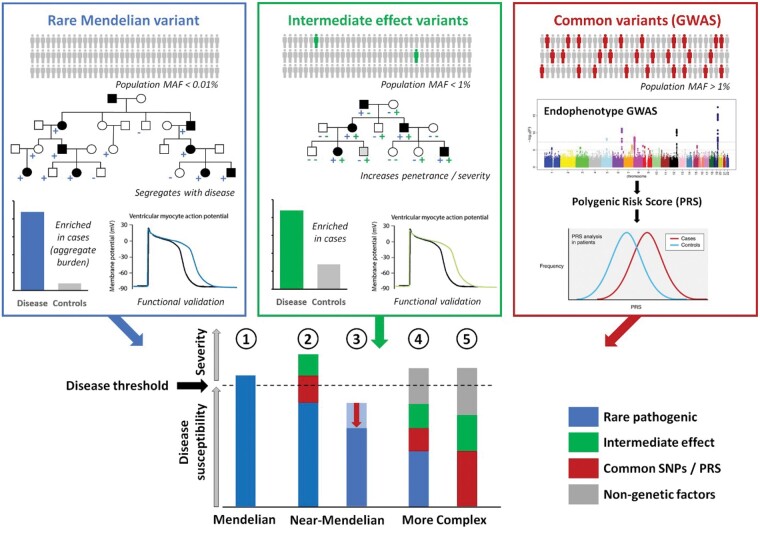
Figure 1.
The genetic aetiology of cardiovascular diseases. Mendelian disease variants (upper left panel) are ultra-rare in the population and have large effect sizes, though often not sufficient in isolation to yield a disease phenotype. Mendelian genes and variants can be identified through analysis of family pedigrees or burden analysis in case–control studies and further validated with functional assays. Common variants (upper right panel) with individually small effect sizes may collectively contribute to disease burden or modulate the effects of Mendelian variants. Intermediate effect variants (upper middle panel) are emerging variant classes that usually have population frequencies and effect sizes between rare Mendelian and common variants and may act to increase severity and penetrance. Such variants can be identified by demonstrating enrichment in case cohorts and deleterious effects in established functional assays. These different variant classes can combine to reach the threshold of disease in patients with rare cardiovascular diseases and contribute to the variable severity observed in patients. Diseases such as HCM and LQTS are often Mendelian [1] or near-Mendelian where Mendelian variants of large effect sizes can combine with other variant classes to cause disease [2] or act as protective modifiers (e.g. regulatory variants affecting the expression ratio of the mutant vs. non-mutant alleles) [3]. In contrast, diseases such as BrS and DCM may exhibit a more complex aetiology where substantial non-Mendelian genetic and non-genetic factors are required to reach disease threshold in the presence of a low penetrance rare variant [4] or in a non-Mendelian disease model [5]. blue −, individual does not harbour the familial rare pathogenic variant; blue +, individual harbours the familial rare pathogenic variant; green −, individual does not harbour that intermediate effect variant; green +, individual harbours a given intermediate effect variant; GWAS, genome-wide association study; MAF, minor allele frequency; PRS, polygenic risk score; SNP, single-nucleotide polymorphism. Adapted from Walsh et al.3