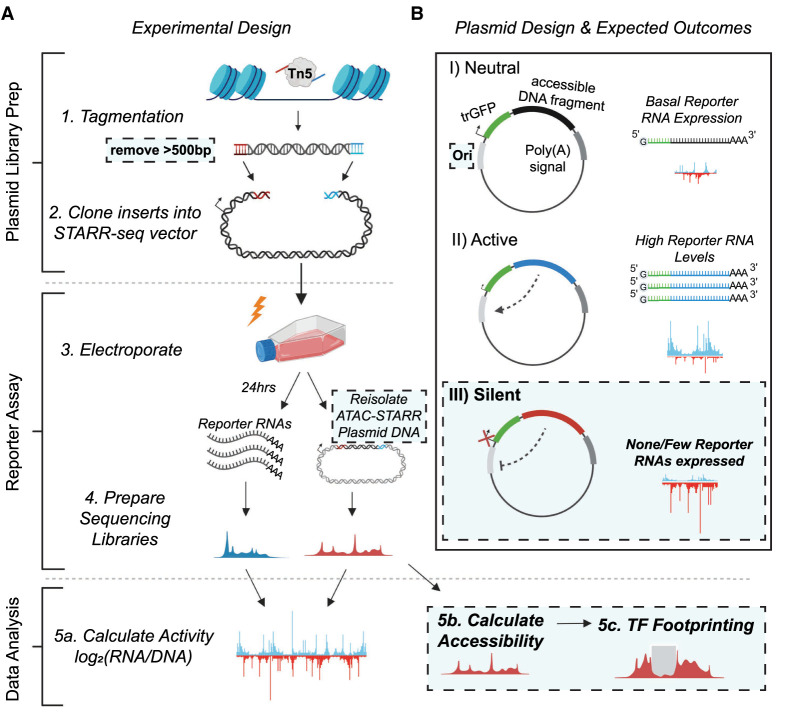
Figure 1.
Schematic of the ATAC-STARR-seq methodology. (A) The experimental design of ATAC-STARR-seq consists of three parts: plasmid library generation; reporter assay; and data analysis. Open chromatin is isolated from cells with the cut-and-paste transposase Tn5 and only large DNA fragments (>500 bp) are removed. The open chromatin fragments are cloned into a reporter plasmid and the resulting clones—called an ATAC-STARR-seq plasmid library—are electroporated into cells. Twenty-four hours later, both reporter RNAs (blue)—which are transcribed directly off the ATAC-STARR-seq plasmid—and ATAC-STARR-seq plasmid DNA (red) are harvested, and Illumina sequencing libraries are prepared and sequenced. The resulting ATAC-STARR-seq data are analyzed to extract regulatory activity, chromatin accessibility, and transcription factor footprints. (B) Reporter plasmid design and the expected outcomes for neutral, active, and silent regulatory elements. Each ATAC-STARR-seq plasmid within a library contains a truncated GFP (trGFP) coding sequence, a polyadenylation signal sequence, an origin of replication (Ori) (which moonlights as a minimal core promoter), and the unique open chromatin fragment being assayed. Because the accessible region is contained in the 3′ UTR, the abundance of itself in the transcript pool reflects its activity. In this way, neutral elements do not affect the system and reporter RNAs are expressed at a basal expression level dictated by the minimal core promoter, the Ori. Accessible chromatin fragments that are active express reporter RNAs at a higher level than the basal expression level, whereas silent elements repress the Ori and reporter RNAs are expressed at a lower level than basal expression. Dashed boxes represent new components of the ATAC-STARR-seq assay design and workflow.