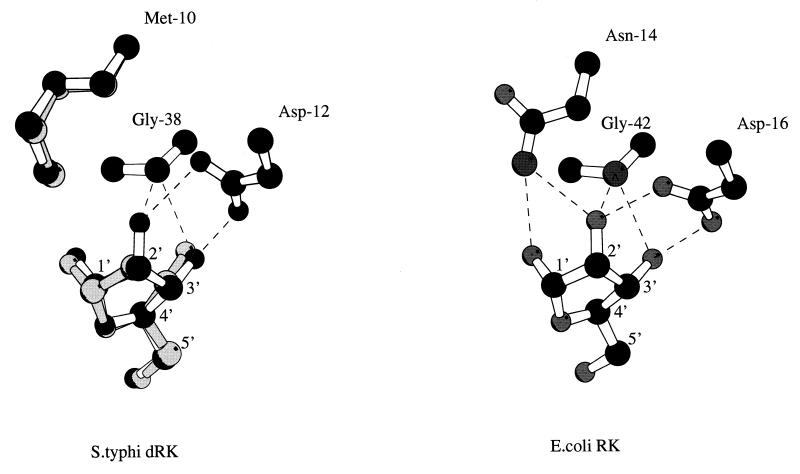
FIG. 5.
Schematic drawing of hydrogen bonding interactions in the substrate binding site. (Left) Superposition of ribose (black) with deoxyribose (gray) and relative positions of equivalent residues in the dRK-ribose and dRK-deoxyribose models. (Right) The three residues involved in the interaction with the ribose O-2′ atom in the E. coli RK (Protein Data Bank code 1rkd) are indicated. Ribose carbon atoms are numbered, and dotted lines correspond to hydrogen bonds. The two complexes were modeled by homology with the X-ray crystal structure of the ternary complex of the E. coli RK with the ribose and ADP (21). The side chain of residues not conserved in the E. coli RK structure was changed using the O program (10), and then the coordinates were energy minimized with the procedure implemented in X-plor (5). The figure was prepared using MOLSCRIPT (11).