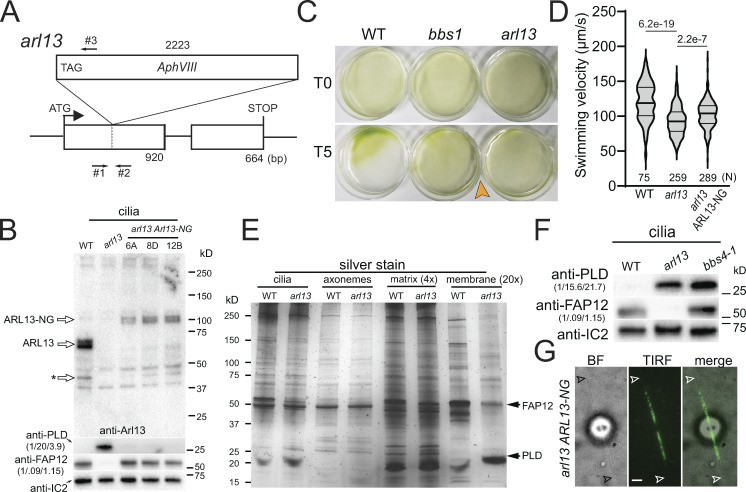
Figure 1.
Chlamydomonas arl13 shows defects in swimming, phototaxis, and ciliary composition. (A) Schematic presentation of ARL13 genomic DNA and the insertion in arl13. The start codon of ARL13 and the positions of the primers used to track the insertion are indicated (arrows #1–3). (B) Western blot of isolated cilia from control (g1, WT), arl13, and three different arl13 ARL13-NG strains, probed with antibodies against ARL13, PLD, FAP12 and, as a loading control, IC2. The star marks a potential ARL13 fragment. The quantification of the band strengths normalized for those of IC2 are shown in brackets and is based on one experiment. See Fig. S2, A and B, for average data based on several biological replicates. (C) Population phototaxis assay of control (g1, WT), bbs1, and arl13. The direction of light and time of exposure in minutes are indicated. (D) Violin plot of the swimming velocities of control (g1, WT), arl13, and arl13 ARL13-NG cells. N, the number of cells analyzed. The P values of a two-tailed t test are indicated. (E) Silver stained SDS-PAGE of isolated cilia and ciliary subfractions from Triton X-114 phase partitioning of the control (g1, WT) and arl13 strains. (F) Western blot comparing isolated cilia from the control (g1, WT), arl13, and bbs4-1 strains, probed with antibodies against PLD, FAP12, and, as a loading control, IC2. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. (G) Bright-field (BF), TIRF, and merged image showing the distribution of ARL13-NG. Arrowheads mark the ciliary tips. Bar = 2 µm. Source data are available for this figure: SourceData F1.