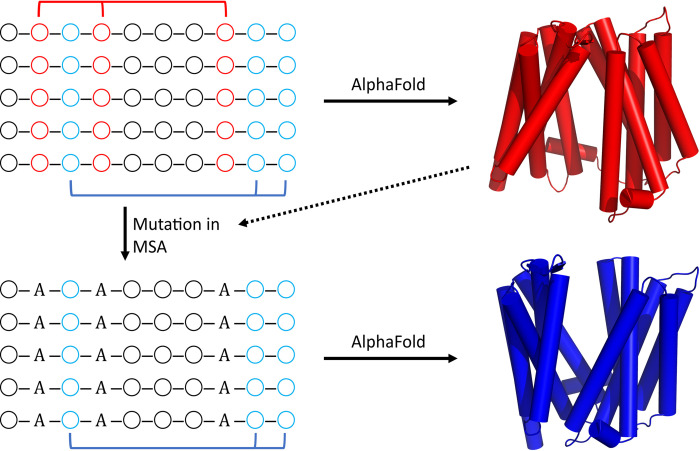
Fig 1. Methodology.
An initially generated MSA, via MMSeqs2, is input into Alphafold2 within ColabFold to generate five structural models. For illustration, the model with the highest pLDDT, AF2’s ranking of model confidence, is shown in red. Residues for mutation are chosen, in this case the three residues in red mediating a contact point on the upper surface of the protein. These mutations are made across the entire MSA (ignoring gaps). This modified MSA is then input into ColabFold for generation of new models. With the contact points in red missing, Alphafold2 within ColabFold generates a new conformation based on the contacts shown in blue.