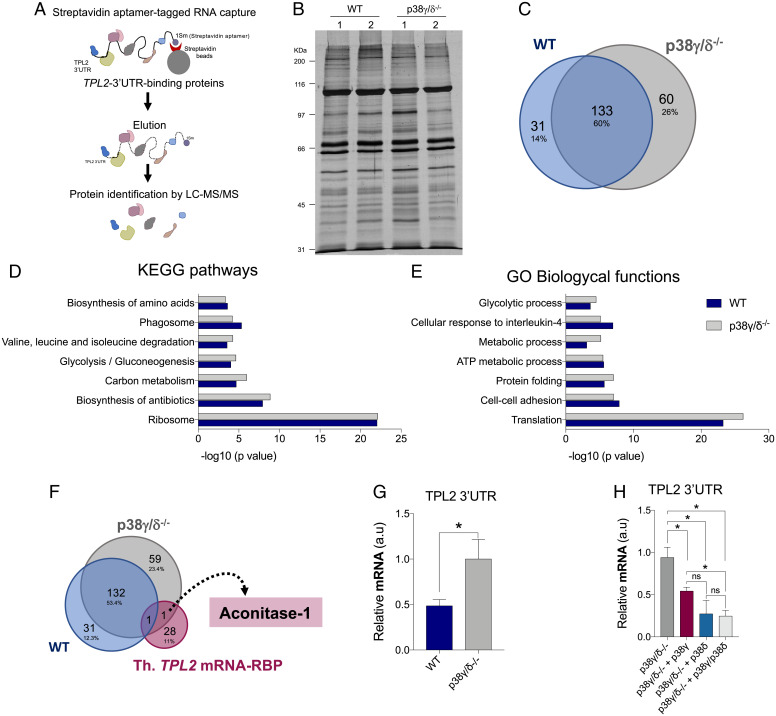
Fig. 4.
Analysis of the proteins bound to the 3′UTR of TPL2 mRNA. (A) Schematic representation of the streptavidin aptamer (1sm)-tagged RNA–protein capture methodology and identification of TPL2 3′UTR-bound proteins. LC-MS/MS, liquid chromatography–tandem mass spectrometry. (B) SDS-PAGE and silver staining of proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− macrophage extracts. Lanes 1 and 2 are biological replicas. (C) Venn diagrams (https://bioinfogp.cnb.csic.es/tools/venny/) showing the number of common and specific proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cell extracts. (D and E) Enrichment analysis of KEGG pathways (D) and GO biological processes (E) of the proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cell extracts. (F) Venn diagrams showing the number of common and specific proteins bound to 3′UTR of TPL2 mRNA from WT and p38γ/δ−/− cells and RBPs containing binding sites in the TPL2 mRNA. Th. TPL2 mRNA-RBP, proteins containing predicted binding sites in TPL2 mRNA 3′UTR. (G) Endogenous ACO1 was immunoprecipitated from WT and p38γ/δ−/− MEFs lysates under Ribonuclease (RNase)-free conditions. RNA was isolated from pellets and retrotranscribed to complementary DNA (cDNA) for qPCR analysis. TPL2 3′UTR RNA levels were quantified and normalized to 18S RNA levels and to each input control. Data show mean ± SEM from two experiments in duplicate. *P ≤ 0.05 relative to p38γ/δ−/− MEFs. (H) p38γ/δ−/− MEFs were transfected with empty HA vector (p38γ/δ−/−) or with plasmids encoding HA-p38γ (p38γ/δ−/− + p38γ), HA-p38δ (p38γ/δ−/− + p38δ), or HA-p38γ plus HA-p38δ (p38γ/δ−/− + p38γ/p38δ), and endogenous ACO1 was immunoprecipitated as described in G. TPL2 3′UTR RNA levels were quantified and normalized to 18S RNA levels and to each input control. Data show mean ± SEM from two experiments in duplicate. ns, not significant. *P ≤ 0.05 relative to p38γ/δ−/− MEFs.