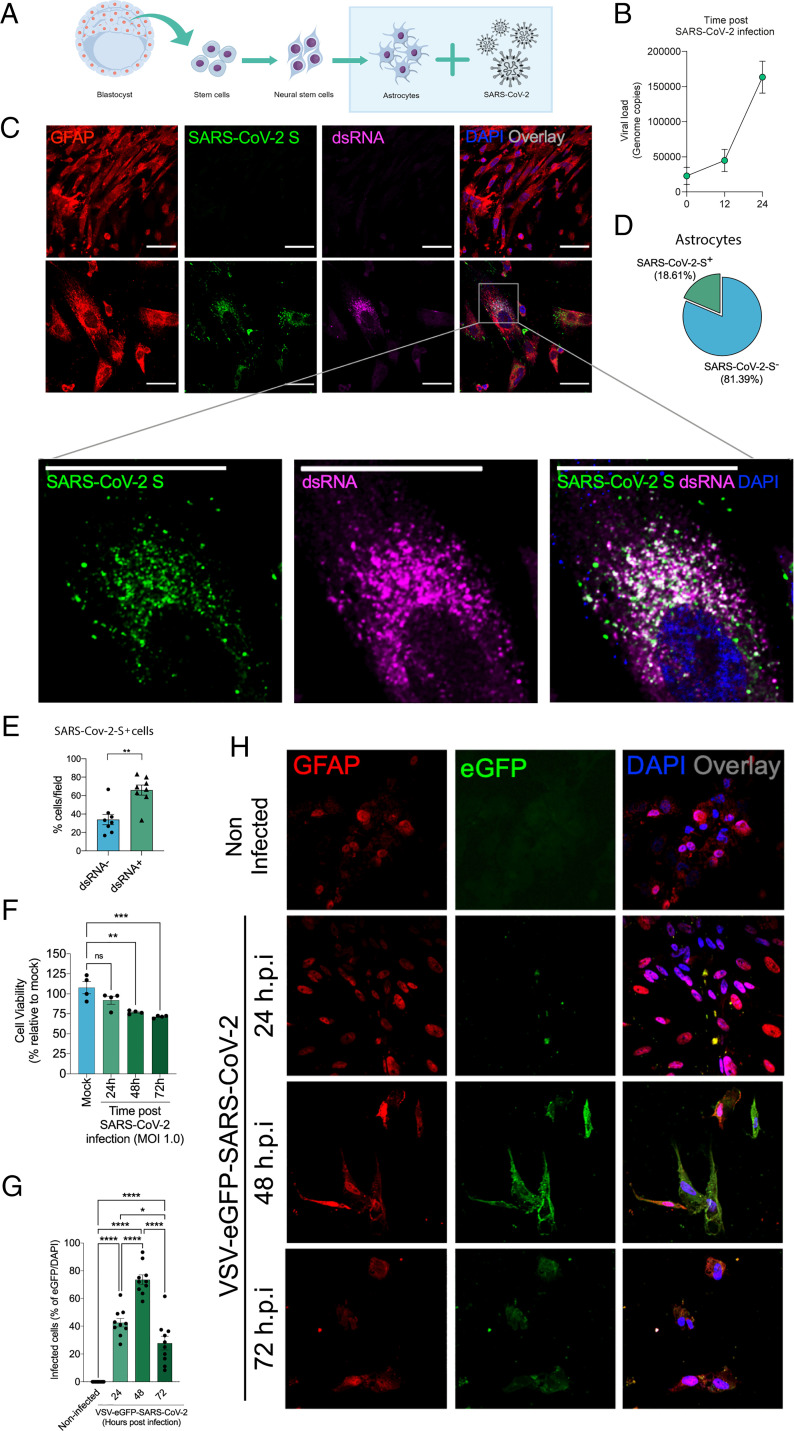
Fig. 3.
SARS-CoV-2 infects and replicates in astrocytes in vitro. (A) Human neural stem cell–derived astrocytes were infected in vitro with SARS-CoV-2 (MOI 1.0) for 1 h, washed, and harvested 24 h after infection. (B) SARS-CoV-2 viral load detection in astrocyte cell pellets (n = 6 replicates) using RT-PCR. (C) Immunostaining for GFAP (red), ds RNA (magenta), SARS-CoV-2-S (green), and nuclei (DAPI; blue). Images were acquired at 630× magnification. (Scale bars: 50 µm.) (D) Percentage of infected astrocytes. The data depict SARS-CoV-2-S and DAPI-stained cells (100 fields were analyzed). (E) Frequency of cells containing replicating viruses. (F) Astrocyte viability upon SARS-CoV-2 infection was assessed using a luminescence-based cell viability assay (CellTiter-Glo), determining the number of live cells by quantification of ATP at 24, 48, and 72 hpi. (G) Percentage of infected cells with pseudotyped SARS-CoV-2 (VSV-eGFP-SARS-CoV-2) at 24, 48, and 72 hpi. (H) Staining for DAPI (nuclei; blue), GFAP (astrocytes; red), and eGFP (virus; green) in astrocytes infected with pseudotyped SARS-CoV-2 (VSV-eGFP-SARS-CoV-2) at 24, 48, and 72 hpi. The data represent the percentage of dsRNA-stained cells of SARS-CoV-2-S–positive cells (10 fields were analyzed). All data are representative of at least two independent experiments performed in triplicate or quadruplicate and shown as mean ± SEM. P values were determined by two-tailed unpaired tests with Welch's correction (E) or one-way ANOVA followed by Tukey’s post hoc test (F and G). *indicate statistical significance. **P < 0.01 compared with the mock group; ***P < 0.001 compared with the mock group; ****P < 0.0001 compared with the mock group. ATP: adenosine triphosphate; MOI: multiplicity of infection; ns: not significant; DAPI: 4′,6-diamidino-2-phenylindole.