Figure 3.
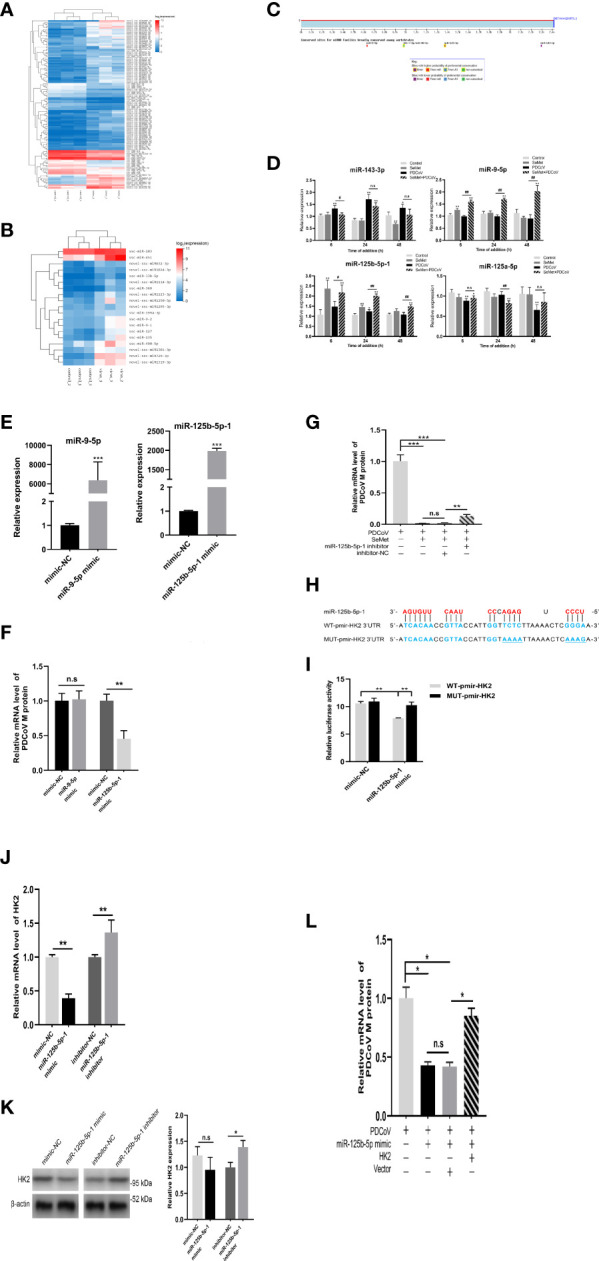
miR-125b-5p-1 is required for HK2-dependent SeMet inhibiting PDCoV replication. (A, B) Hierarchic clustering analyses of 118 miRNAs that were differentially expressed in the SeMet-supplemented LLC-PK1 cells by >2-fold compared with the mock-treated LLC-PK1 cells, and 19 miRNAs that were differentially expressed in the PDCoV-infected cells by >2-fold compared with the mock-treated cells. (C) Analysis of the TargetScanHuman (http://www.targetscan.org/vert_72/) website shows that miR-143-3p, miR-9-5p, miR-125b-5p-1, and miR-125a-5p can target the human 3’UTR of HK2. Binding sequences are highly conserved among species (including human and swine). (D) LLC-PK1 cells were treated with PDCoV (100 TCID50) and SeMet (150 μM) for 6, 24, and 48 h, and the levels of miR-143-3p, miR-9-5p, miR-125b-5p-1, and miR-125a-5p were determined by qRT-PCR (n=6). (E) The miR-9-5p and miR-125b-5p-1 expressions were determined by qRT-PCR when transfected with mimic-NC, miR-9-5p mimic, and miR-125b-5p-1 mimic (n=4). (F) Viral mRNA in the LLC-PK1 cells transfected with miR-9-5p mimic or miR-125b-5p-1 mimic was quantified by qRT-PCR after 48 h (n=6-8). (G) Cells were transfected with miR-125b-5p-1 inhibitor, followed by treatment with SeMet and PDCoV. Viral mRNA expression was analyzed by qRT-PCR (n=6). (H) The miR-125b-5p-1 targets the base sequence and mutant sequence of the 3’UTR of HK2. (I) The miR-125b-5p-1 mimic or mimic-NC and WT-pmirGLO-HK2 or MUT-pmirGLO-HK2 were co-transfected into HEK293T cells. The luciferase reporter assay was used to detect whether miR-125b-5p-1 targeted to bind to the 3’UTR of HK2 (n=5). (J, K) The HK2 mRNA and protein expressions were determined by qRT-PCR and Western blot in LLC-PK1 cells when transfected with miR-125b-5p-1 mimic or inhibitor (n=3-6). (L) LLC-PK1 cells were transfected with miR-125b-5p-1 mimic and pcDNA3.1-HK2, followed by infection with PDCoV. Viral mRNA expression was analyzed by qRT-PCR (n=6). Means ± SD are shown. Statistical significiance was determined by Student t test. ***, P<0.001; **, P<0.01; *, P<0.05; ##, P<0.01; #, P<0.05; n.s: not significant. All experiments were repeated at least twice and representative results are shown.
