FIG. 2.
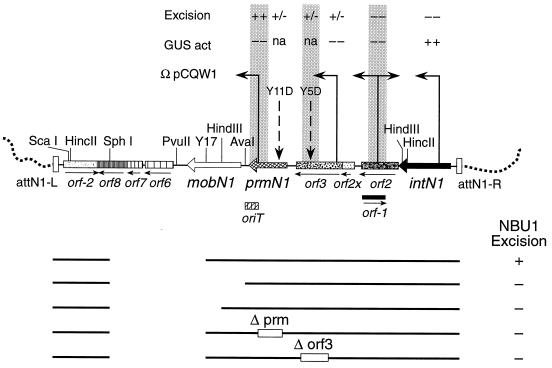
Regions of NBU1 required for excision. NBU1 is shown integrated in the Bacteroides target site. The chromosomal Leu-tRNA gene in the chromosomal target site is located upstream of attN-L. The possible ORFs and known genes are labeled, and the locations of the integrated forms of pEG920 insertion vectors, pY5D in orf3, pY11D in prmN1, and pY17 in mobN1, are indicated. The insertion sites and orientations of the uidA reporter vector, pCQW1, are indicated by the arrows above the map. The insertions were tested for their effect on the excision of NBU1 and the level of transcription of the ORFs as measured by the production of GUS from the pCQW1 insertions. Excision was determined by the detection of the NBU1 joined ends on Southern blots, and the results are expressed relative to the intensity observed for wild-type NBU1: wild excision (++) and weak but detectable excision (+/−). Transcription levels of the various pCQW insertions are relative to the GUS activity for the insertion in intN1: 7 U/mg of protein (++) and <0.2 U/mg of protein (−). Transcription could not be measured for the pY5D and pY11D insertions (not applicable [na]). The subcloned region of NBU1 required for both integration and excision on pNW18 and subcloned on a 7.7-kbp SphI fragment into pGERM, pG-Sph18, is indicated below the map. None of the ORFs between PvuII and attN-L are required for either integration or for excision. Other NBU1 subclones of the 7.7-kbp SphI fragment in pGERM used to determine the region required for excision are also shown: SphI-AvaI, Sph-HindIII, and pG-Sph18 with deletions in prmN1 (Δprm, pG-Sph18Δprm) and orf3 (Δorf3, pG-Sph18Δorf3). The 220-bp oriT, required in cis for mobilization, is also part of the XRS between prmN1 and Y17 that is required in cis for NBU1 excision. The excision (+) or lack of excision (−) of each subclone is shown to the right.