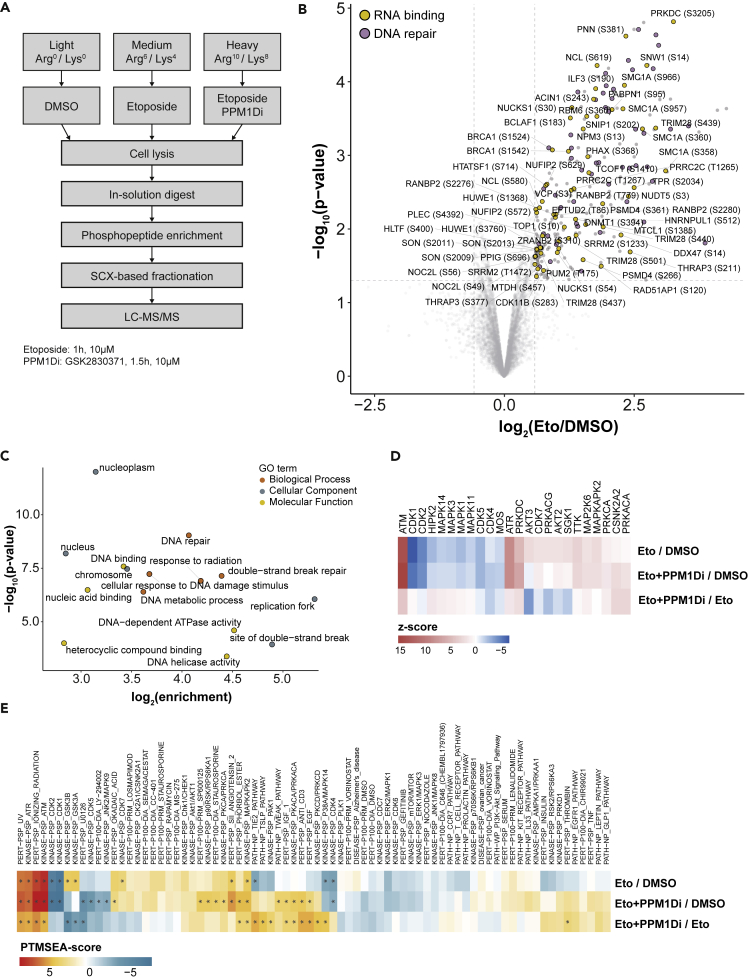
Figure 1.
Phosphoproteomic analysis of the etoposide-induced and PPM1D-dependent DNA damage response
(A) Schematic representation of the strategy used for phosphoproteomic analysis. Light-, medium- or heavy-labeled U2OS cells were treated either with DMSO, with 10μM etoposide for 1h or with 10μM PPM1D inhibitor for 1.5h followed by etoposide treatment. Cells were lysed and digested using trypsin followed by TiO2-based phosphopeptide enrichment and LC-MS/MS analysis. The experiment was performed in triplicates.
(B) Volcano plot showing upregulated phosphorylation sites after etoposide treatment (fold change (FC) > 1.5, moderated t-test: p value < 0.05). Phosphorylation sites on proteins mapping to the GO terms DNA repair and RNA binding are highlighted and RNA binding proteins are labeled. Phosphorylation sites with an FC below −2.5 are not shown.
(C) GO term analysis of upregulated phosphorylation sites after etoposide treatment using ViseaGO R package (Fisher exact test: p value < 0.05).
(D) Kinase-substrate enrichment analysis of etoposide-induced and PPM1D-dependent phosphorylation sites. Relative Z score indicates changes in kinase activities after indicated treatments (One-tailed probability test: p value < 0.05).
(E) PTM set enrichment analysis showing phosphorylation site-specific pathways, perturbations, and kinase activities (Kolmogorov-Smirnov test: ∗ Benjamini-Hochberg adj. p value < 0.05).