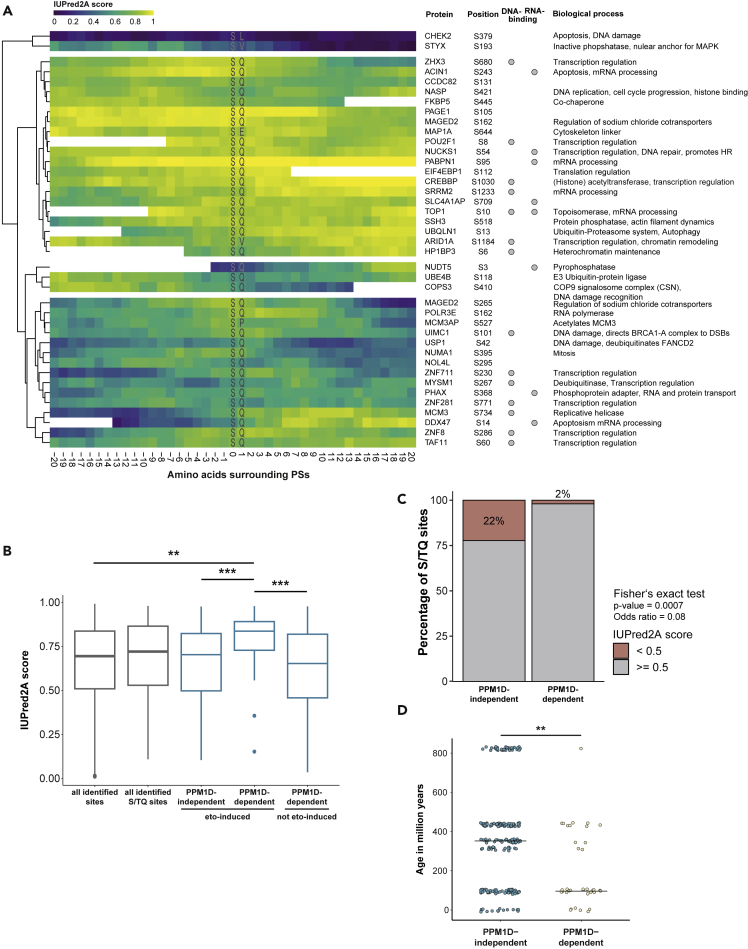
Figure 4.
DNA damage-induced PPM1D substrates are located in intrinsically disordered protein regions
(A) Intrinsic disorder score (IUPred2A) of surrounding protein regions of etoposide- and PPM1D-dependent phosphorylation sites. IUPred2A score above 0.5 is considered as disorder. Known DNA- and RNA-binding motifs and biological processes of phosphorylated proteins are annotated.
(B) Comparison of IUPred2A score of different subsets of etoposide- and PPM1D-targeted sites with S/TQ-motif sites and all identified phosphorylation sites from the phosphoproteome (T-test: ∗∗ p value < 0.001, ∗∗∗ p value < 0.0001).
(C) Barplot showing the fraction of S/TQ sites with IUPred2A score <0.5 (not disordered) within PPM1D-dependent sites (upregulated in H/M condition) and PPM1D-independent sites (not upregulated in H/M condition) regardless of their regulation status after etoposide treatment. Fisher's exact test was carried out on the contingency table of S/TQ site counts in each subset.
(D) Comparison of estimated phosphorylation site age (+/− 3 amino acids) based on ptmAge prediction of etoposide-induced and PPM1Di-responsive sites with etoposide-induced and PPM1D-independent sites (Cochran-Armitage trend test: ∗∗ p value < 0.001). Datapoints are jittered.