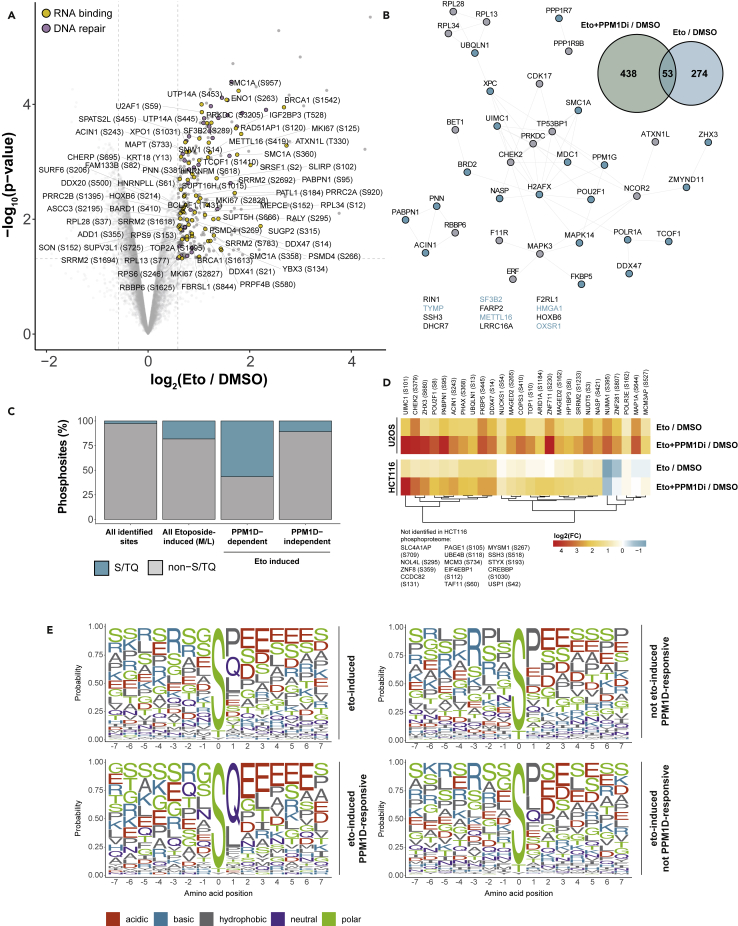
Figure 5.
Phosphoproteomic analysis of PPM1D substrates after etoposide treatment in HCT116 cells
(A) Volcano plot of phosphorylation sites after etoposide treatment (FC > 1.5, moderated t-test: p value < 0.05). Phosphorylation sites on proteins mapping to the GO terms DNA Repair and RNA binding are highlighted and RNA binding proteins are labeled.
(B) Etoposide-induced site was overlapped with upregulated sites after combined etoposide and PPM1Di treatment compared to etoposide treatment. Network showing proteins containing the 53 overlapping phosphorylation sites (STRING conf. score >0.4). Sites containing an SQ motif are colored in blue and proteins without any known interaction partner in the network are shown below.
(C) Fractions (%) of S/TQ motif abundance in all identified sites compared to the etoposide-induced, PPM1D-dependent, and PPM1D-independent subset.
(D) Log2 fold changes in HCT116 screen of etoposide- and PPM1Di-induced phosphorylation sites from the U2OS screen. Sites that are not identified in the HCT116 screen are annotated aside.
(E) Sequence motif analysis of sites belonging to the different subsets from the HCT116 screen. Amino acid probabilities are plotted using the ggseqlogo R package.