Figure 6.
Kanadaptin is phosphorylated by ATM and dephosphorylated by PPM1D in response to DSBs
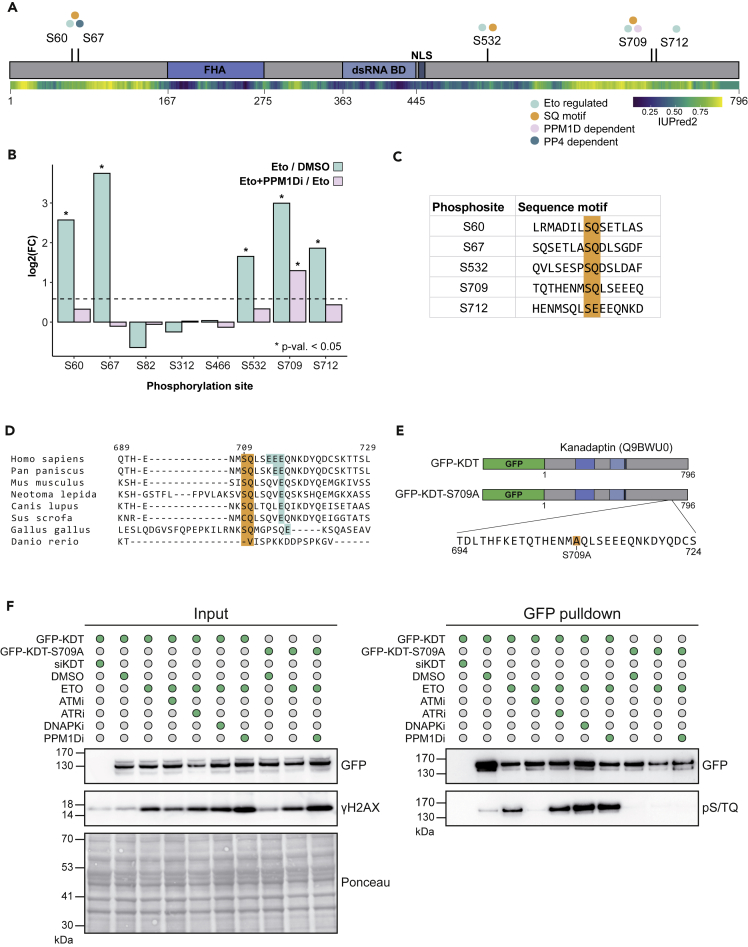
(A) Scheme of human Kanadaptin (Q9BWU0) containing an FHA domain, SMART-predicted double-stranded RNA binding domain, and nuclear localization signal (NLS). Regulated phosphorylation sites after etoposide and PPM1Di treatment are annotated with their dependencies and SQ sites are indicated. Regulation by PP4 is predicted based on (Ueki et al., 2019) IUPred2A score is mapped to the protein and represents intrinsic disorder.
(B) log2-transformed FCs of all Kanadaptin phosphorylation sites identified by phosphoproteomics (moderated t-test: ∗ p value < 0.05).
(C) Amino acid environment of significantly upregulated Kanadaptin phosphorylation sites after DNA damage.
(D) Multiple sequence alignment (ClustalW) of pS709/S712 motif. Phosphorylated serine +1 amino acid and downstream glutamic acid residues are highlighted.
(E) Scheme of GFP tagged wt-Kanadaptin and phospho-dead (S709A) constructs.
(F) GFP-Kanadaptin or GFP-Kanadaptin-S709A were transiently expressed in U2OS cells treated with 10μM etoposide for 1h, or additionally treated with ATMi (10μM), ATRi (2μM), PPM1Di (10μM), or DNAPKi (10μM) for 1.5h, or co-transfected with siRNA against Kanadaptin. GFP-Kanadaptin was pulled down followed by washes in 8M urea.