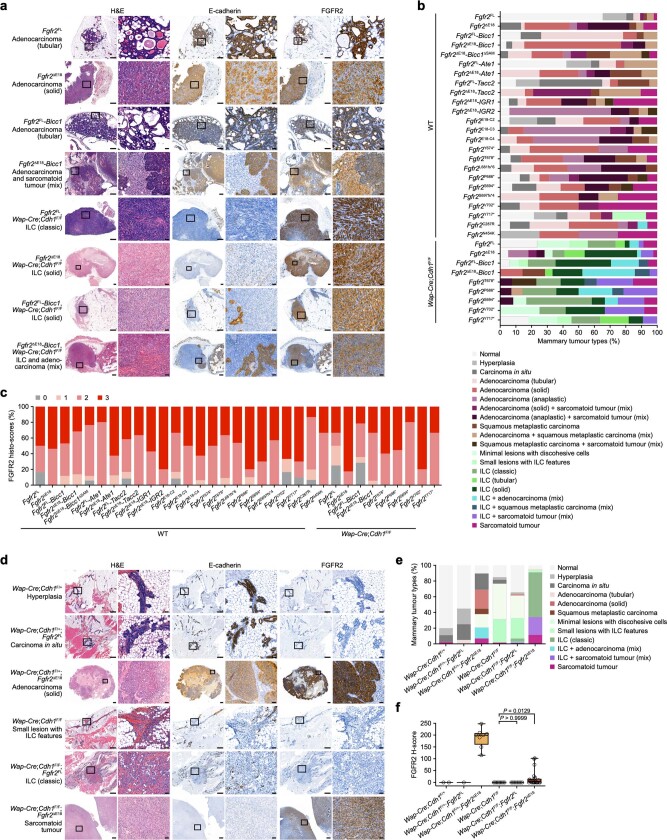
Extended Data Fig. 8. Mammary tumour types observed in Fgfr2 mouse models.
a, Representative hematoxylin and eosin (H&E) histochemistry and FGFR2 and E-cadherin immunohistochemistry (IHC) stains on mammary tissue sections from indicated Fgfr2 somatic mouse models. Per MG one tissue section was stained and quantified for each of the indicated stains acquired in multiple independent randomized batches across all Fgfr2 variants. Numbers of stained and quantified MGs are in (b, c). ILC, invasive lobular carcinoma. b, Mammary tumour type classifications of Fgfr2 somatic mouse models based on H&Es and E-cadherin IHC stains. WT, Fgfr2FL, n = 46 of 20; Fgfr2ΔE18, n = 45 of 22; Fgfr2FL-Bicc1, n = 23 of 10; Fgfr2ΔE18-Bicc1, n = 26 of 11; Fgfr2ΔE18-Bicc1ΔSAM, Fgfr2ΔE18-Ate1, Fgfr2FL-Tacc2, Fgfr2E18-C3, n = 20 of 10; Fgfr2FL-Ate1, Fgfr2ΔE18-Tacc2, Fgfr2E18-C4, n = 19 of 10; Fgfr2ΔE18-IGR1, n = 18 of 9; Fgfr2ΔE18-IGR2, Fgfr2E18-C2, n = 17 of 10; Fgfr2Y674*, Fgfr2C287R, n = 16 of 8; Fgfr2T678*, n = 15 of 4; Fgfr2L681fs*6, n = 14 of 7; Fgfr2P686*, n = 16 of 5; Fgfr2S694*, n = 13 of 5; Fgfr2S697fs*4, n = 8 of 4; Fgfr2V702*, n = 8 of 3; Fgfr2Y717*, n = 14 of 5; Fgfr2N454K, n = 4 injected MGs of 2 mice. Wap-Cre;Cdh1F/F, Fgfr2FL, n = 34 of 15; Fgfr2ΔE18, n = 39 of 15; Fgfr2FL-Bicc1, n = 17 of 9; Fgfr2ΔE18-Bicc1, n = 21 of 11; Fgfr2T678*, n = 13 of 7; Fgfr2P686*, n = 14 of 4; Fgfr2S694*, n = 12 of 4; Fgfr2V702*, n = 12 of 5; Fgfr2Y717*, n = 11 injected MGs of 4 mice. c, Histo-scoring of FGFR2 IHC stains on mammary tumours from Fgfr2 somatic mouse models. WT, Fgfr2FL, n = 6 of 5; Fgfr2ΔE18, n = 39 of 22; Fgfr2FL-Bicc1, n = 17 of 10; Fgfr2ΔE18-Bicc1, n = 22 of 11; Fgfr2ΔE18-Bicc1ΔSAM, n = 17 of 9; Fgfr2FL-Ate1, n = 5 of 5; Fgfr2ΔE18-Ate1, Fgfr2E18-C4, n = 16 of 9; Fgfr2FL-Tacc2, n = 12 of 8; Fgfr2ΔE18-Tacc2, n = 11 of 8; Fgfr2ΔE18-IGR1, Fgfr2E18-C3, n = 14 of 9; Fgfr2ΔE18-IGR2, n = 15 of 10; Fgfr2E18-C2, n = 12 of 9; Fgfr2Y674*, n = 14 of 8; Fgfr2T678*, n = 11 of 4; Fgfr2L681fs*6, n = 13 of 8; Fgfr2P686*, Fgfr2S694*, n = 10 of 5; Fgfr2S697fs*4, n = 7 of 4; Fgfr2V702*, n = 6 of 3; Fgfr2Y717*, n = 10 of 4; Fgfr2C287R, n = 15 of 8; Fgfr2N454K, n = 3 tumours of 2 mice. Wap-Cre;Cdh1F/F, Fgfr2FL, n = 12 of 8; Fgfr2ΔE18, n = 23 of 9; Fgfr2FL-Bicc1, n = 14 of 7; Fgfr2ΔE18-Bicc1, n = 18 of 10; Fgfr2T678*, Fgfr2V702*, n = 10 of 5; Fgfr2P686*, n = 9 of 4; Fgfr2S694*, n = 10 of 4; Fgfr2Y717*, n = 6 tumours of 3 mice. d, Representative H&E histochemistry and E-cadherin and FGFR2 IHC stains on mammary tissue sections from indicated GEMMs. Per MG one tissue section was stained and quantified for each of the indicated stains acquired in two independent randomized batches across all genotypes. Numbers of stained and quantified MGs are in (e, f). e, Mammary tumour type classifications of GEMMs based on H&Es and E-cadherin IHC stains. Wap-Cre;Cdh1F/+, n = 45 of 12; Wap-Cre;Cdh1F/+;Fgfr2FL, n = 20 of 5; Wap-Cre;Cdh1F/+;Fgfr2ΔE18, n = 29 of 6; Wap-Cre;Cdh1F/F, n = 60 of 16; Wap-Cre;Cdh1F/F;Fgfr2FL, n = 73 of 19; Wap-Cre;Cdh1F/F;Fgfr2ΔE18, n = 79 MGs of 19 mice. f, Histo (H)-score quantifications of FGFR2 IHC stains on mammary tumours from GEMMs. Wap-Cre;Cdh1F/+, n = 2 of 2; Wap-Cre;Cdh1F/+;Fgfr2FL, n = 1 of 1; Wap-Cre;Cdh1F/+;Fgfr2ΔE18, n = 8 of 5; Wap-Cre;Cdh1F/F, n = 9 of 8; Wap-Cre;Cdh1F/F;Fgfr2FL, n = 8 of 6; Wap-Cre;Cdh1F/F;Fgfr2ΔE18, n = 24 tumours of 19 mice. Data are represented as median (centre line) ± IQR (25th to 75th percentile, box) and ± full range (minimum to maximum, whiskers) and P values were calculated with one-tailed Kruskal-Wallis tests and Dunn’s multiple-testing corrections. Scale bars, overview, 500 μm; inset, 50 μm.