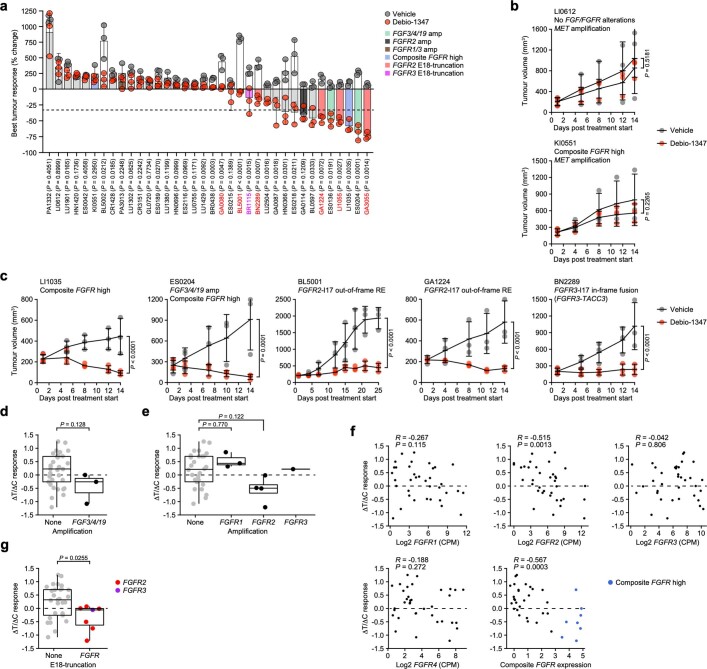
Extended Data Fig. 15. PDXs expressing FGFR2ΔE18 variants are sensitive to debio-1347.
a, Best percentage change from baseline tumour volume in patient-derived xenograft (PDX) models (n = 36) engrafted in female NOD-SCID (BL5001, BL5002) or BALB/c Nude (all other PDX models) mice and treated daily orally with vehicle or debio-1347 (n = 3 mice per PDX model and treatment group). Coloured bars indicate identified FGF/FGFR2 alterations detailed in Fig. 4c. b, c, Growth curves of indicated PDXs models engrafted in NOD-SCID or BALB/c Nude mice and treated daily orally with vehicle or debio-1347 (ES0204, Li0612, LI1035, BN2289, 60 mg/kg; GA1224, KI0551, 80 mg/kg; BL5001, day 1-14, 40 mg/kg; day 15-25, 60 mg/kg; n = 3 mice per PDX model and treatment group). d, Debio-1347 ΔT/ΔC response ratios in PDXs with FGF3/4/19 amp (n = 3) versus normal CN (n = 33). e, Debio-1347 ΔT/ΔC response ratios in PDXs with FGFR1 amp (n = 3), FGFR2 amp (n = 4), or FGFR3 amp (n = 1) versus normal CN (n = 28). f, Correlations of FGFR1, FGFR2, FGFR3, FGFR4, or composite FGFR expression versus debio-1347 ΔT/ΔC response ratios across PDXs. Composite FGFR expression was defined as high, if normalized expression > 3. g, Debio-1347 ΔT/ΔC response ratios in PDXs expressing E18-truncated FGFR2 (n = 6) or FGFR3 (n = 1) versus no truncation (n = 29). Data are represented as mean ± s.d. (a, b) or as median (centre line) ± IQR (25th to 75th percentile, box) and IQR ± 1.5 x IQR (whiskers) (d, f). P values were calculated with two-tailed unpaired Student’s t-tests (a), one-tailed two-way ANOVA and FDR multiple-testing corrections using the two-stage step-up method from Benjamini, Krieger, and Yekutieli (b, c), two-tailed Wilcoxon rank-sum tests (d, g), one-tailed one-way ANOVA and Tukey’s multiple-testing corrections (e), or two-tailed t-transformations of Pearson’s R correlation coefficients (f).