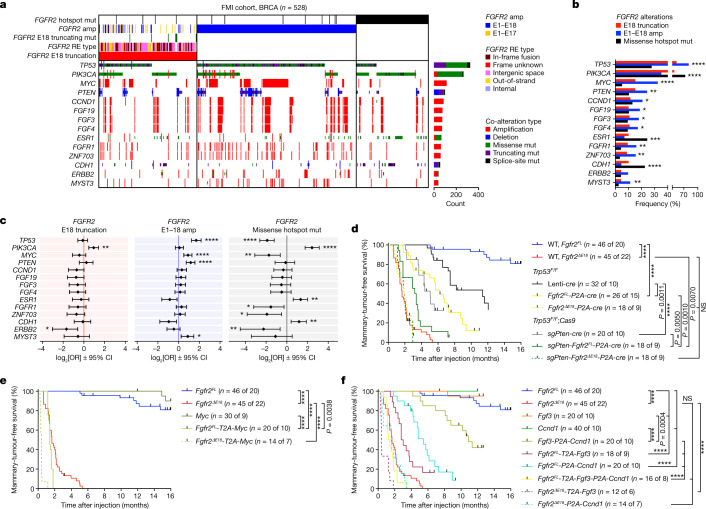
Fig. 3. The oncogenic competence of FGFR2 alterations depends on co-occurring drivers.
a, Analysis of 528 breast cancer samples classified as either FGFR2 E18-truncated (n = 157, 29.7% of total, 0.70% incidence), E1–E18 amplified (n = 256, 48.5% of total, 1.14% incidence) or missense hotspot mutant (n = 115, 21.8% of total, 0.51% incidence), and the top co-enriched tumour driver alterations found in 22,380 breast cancer profiles from FMI. b, Enrichment of the top tumour driver co-alterations in the indicated FGFR2 alteration categories in the FMI breast cancer cohort. c, The odds ratios (ORs) of the top tumour driver co-alterations in the indicated FGFR2 alteration categories (E18 truncation, n = 157; E1–E18 amplification, n = 256; missense hotspot mutation, n = 115) versus FGFR2 WT samples (n = 22,307) of the FMI breast cancer cohort. Data are represented as log2-transformed OR ± 95% confidence interval (CI). Co-occurrence, OR > 1; mutual exclusivity, OR < 1. P values were calculated using one-tailed proportion Z-tests (b) or two-tailed Fisher’s exact tests (c) with FDR multiple-testing corrections using the Benjamini–Hochberg method (b and c). Sample sizes and statistical details for b and c are shown in Supplementary Table 2. d–f, Kaplan–Meier analysis of the mammary-tumour-free survival of Trp53F/F and Trp53F/F;Rosa26-Cas9 (d) or WT (e,f) female mice that were intraductally injected with lentiviruses encoding the indicated variants. Cohort counts (n) represent injected mammary glands (MGs) per number of mice. The Fgfr2FL and Fgfr2ΔE18 curves in d–f are duplicates from Fig. 2c. P values were calculated using log-rank (Mantel–Cox) tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.