Abstract
Secretion systems utilize ATPase activity to facilitate the translocation of proteins into and across membranes. In bacteria, the universally conserved SecA ATPase binds a large repertoire of preproteins and interacts with the SecYEG translocon. In contrast, the type 7b secretion system (T7bSS) of Staphylococcus aureus supports the secretion of a restricted subset of proteins. T7bSSs are found in several Firmicutes as gene clusters encoding secreted WXG100 proteins and FtsK/SpoIIIE-like ATPase. In S. aureus, this ATPase is called EssC and comprises two cytosolic forkhead-associated domains (FHA1–2), two membrane-spanning segments (TM1–2), and four cytosolic modules named DUF (domain of unknown function) and ATPases1-3 (D1D2D3). However, a detailed understanding of the interactions of EssC in the T7bSS is not clear. Here, we tagged EssC and performed affinity chromatography of detergent-solubilized extracts of wild type and isogenic mutants of S. aureus. We found that EssC recruits EsaA, EssA, and EssB in a complex referred to as the ESS (ESAT-6 like secretion system) translocon, and secreted substrates were not required for translocon assembly. Furthermore, deletions of FHA1 and DUF rendered EssC unstable, whereas FHA2 was required for association with EssB. This interaction was independent of EsaA, but EsaA was required to recruit EssA to the EssC–EssB complex. Finally, we show that assembly of the ESS translocon was impaired upon mutation of D2 structural motifs. Together, our data indicate that the ESS translocon is maintained fully assembled at the plasma membrane and that D2 is fundamental in sustaining the integrity of this complex.
Keywords: protein secretion, protein complex, membrane protein, ATPases associated with diverse cellular activities, Staphylococcus aureus, Gram-positive bacteria
Abbreviations: AAA, ATPase Associated with various cellular Activities; DDM, n-dodecyl β-d-maltoside; DUF, domain of unknown function; ESS, ESAT-6 like secretion system; FHA, forkhead-associated domain; T7bSS, type 7b secretion system; TS, Twin-Strep
Protein secretion across membranes is facilitated by channel-forming proteins and often powered by ATP or the proton motive force. Dedicated secretion systems control the selective entry of proteins into cognate secretion channels as well as the exit site where substrates are released (1, 2). The final location of secreted substrates and the number of lipid layers crossed during this process account for the diversity and complexity of secretion machinery. For example, in bacteria, preproteins recognized by the SecA ATPase are delivered to the membrane-embedded SecYEG translocon (3). In the absence of additional topogenic or retention sequences, SecA substrates end up in the periplasm of Gram-negative bacteria or the extracellular milieu of Gram-positive bacterial cultures. Although the repertoire of SecA substrates is extensive and highly variable, translocation is highly efficient and preproteins do not accumulate in the cytosol. This is unlike secretion mediated by the highly specialized type 7b secretion system (T7bSS). In Staphylococcus aureus, the T7bSS is encoded by the ESS (ESAT-6 like secretion system) gene cluster, which encompasses both transport and secreted proteins (4). T7bSSs are found in several families of Firmicutes, including Bacillaceae, Listeriaceae, Staphylococcaceae, and Clostridiaceae, and are defined by the presence of an ATPase that belongs to the FtsK/SpoIIIE family of proteins and at least one small protein of approximately 100 amino acids with the central Trp-X-Gly motif (WXG100 domain) (5, 6). The association of WXG100 and FtsK/SpoIIIE family of proteins was first described based on a comparative genomics study of Firmicutes and Actinomyces (5) to account for the extracellular location of Mycobacterium tuberculosis WXG100 proteins, ESAT-6 and CFP-10 (7). ESAT-6 and CFP-10 are encoded by the adjacent esxA and esxB genes within the ESX-1 gene cluster, the first of five related clusters ultimately defined as Sec-independent secretion systems in mycobacterial species (8). The remaining genes within ESX clusters of Actinomyces and ESS clusters of Firmicutes are not conserved (4, 5, 9, 10, 11). Further ESX clusters have expanded their secretion repertoire to the PE/PPE family of proteins, whereas ESS clusters secrete proteins of the LXG family (12, 13). The two secretion systems are thus referred as T7a and T7b, respectively (8). Most S. aureus strains encode two canonical WXG100 proteins, EsxA and EsxB, and two WXG100-like proteins, EsxC and EsxD (14, 15, 16). The current model is that each protein folds into a hairpin of two antiparallel α-helices bent around the WXG motif (when present) and associates as homodimers or heterodimers resulting in tetrahelical bundles (15, 17, 18, 19). Unlike Sec-dependent substrates, WXG100 proteins are not processed during secretion, and folded dimers accumulate in the cytosol. WXG100 proteins are encountered in both pathogenic and environmental species of bacteria. In S. aureus, mutants unable to perform T7b secretion are less virulent in the mouse model of bloodstream infection, yet the exact role of T7b during infection remains unknown (4, 20). T7 secretion of larger proteins with an N-terminal WXG100-like domain was first reported in Bacillus anthracis (21). Bioinformatic analyses redefined this class of substrates as the LXG family of polymorphic toxins (PFAM: PF04740) (22) and identified a genomic association with immunity factors that were postulated to prevent self-intoxication by cognate polymorphic toxins (22, 23). Multiple copies of seemingly redundant immunity proteins have been proposed to protect from LXG-mediated antagonism between closely related strains (22, 24). Two such proteins, EssD (EsaD) and TspA, were identified in the culture medium of S. aureus (25, 26, 27, 28, 29) and have been proposed to mediate intraspecies competition in vitro (28, 29). The latter implies translocation of polymorphic toxins across the envelopes of both attacking and prey cells by a mechanism not fully understood (10, 11). Nevertheless, in the attacking cell, EssD (EsaD) interacts tightly with its immunity factor EssI (EsaG) and proposed chaperone EssE (EsaE) (25, 26, 27, 28).
Earlier studies found that EssB, a protein with one transmembrane domain, copurified with three other membrane proteins of the ESS cluster, EsaA, EssA, and EssC (30). Here, we tagged EssC to further characterize the ESS/T7b translocon. We selected EssC because it is conserved with the T7a ATPase EccC, which has been shown to oligomerize and bind WXG100 substrates. Furthermore, soluble domains of T7b EssC have been crystallized from various Firmicutes, including S. aureus and Geobacillus thermodenitrificans, and can be compared with partial X-ray structures of Thermomonospora curvata and Mycobacterium smegmatis EccC homologs (31, 32, 33, 34, 35). EssC has also been proposed to promote substrate selection and translocation (36, 37). After generating and purifying several variants of EssC, we report that the N-terminal FHA2 (forkhead-associated domain) and C-terminal D2 domains of EssC contribute to the assembly and stability of the ESS translocon. We also find that the translocon assembles in the absence of the secreted substrates encoded within the ESS cluster.
Results
Purification and composition of the T7b–ESS membrane complex
In S. aureus strain USA300 FPR3727 (38), herein used as the wildtype strain, the essC gene encodes a protein of 1479 amino acids. Two hydrophobic segments, Ile236–Val252 and Ile255–Ile274, span the plasma membrane twice, positioning the N and C termini of EssC in the cytosol; only a very small loop faces the exterior of the cell (Fig. 1, A and B). X-ray crystallography studies revealed that the soluble N-terminal segment folds into two distinct FHA domains comprised of 9 and 12 stacked β-sheets, respectively (31). The cytosolic C-terminal sequence encompasses the domain of unknown function (DUF) followed by three subdomains with characteristic ATPase-like folds. Herein, EssC subdomains are designated FHA1–2, TM1–2, DUF, and D1D2D3, respectively (Fig. 1, A and B). D1 and D2 share the greatest amino acid conservation amongst EssC homologs of Firmicutes (Fig. S1A). FHA domains are conspicuously missing in most strains of B. anthracis (Fig. S1A) and are absent in the five EccC proteins (EccC1–5) of M. tuberculosis H37Rv (Fig. S1B). Overall, the sequences of EccC proteins are clearly distinct from that of S. aureus EssC (Fig. S1, B and C).
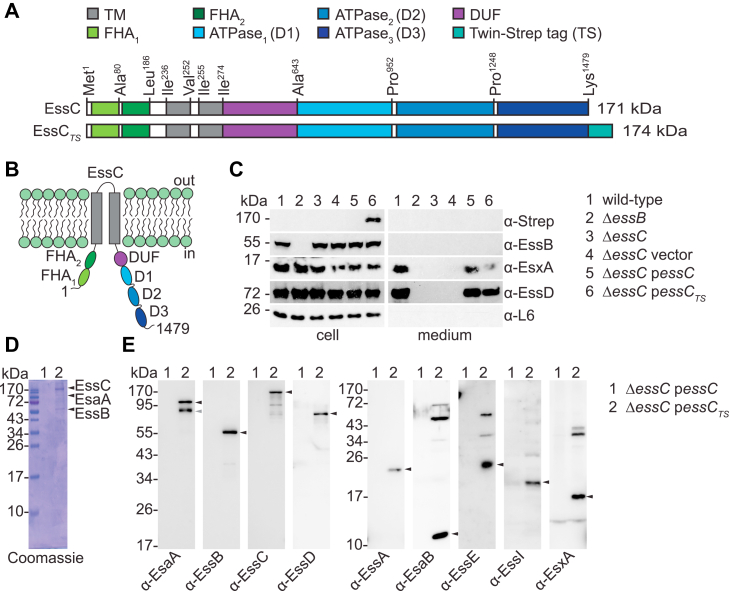
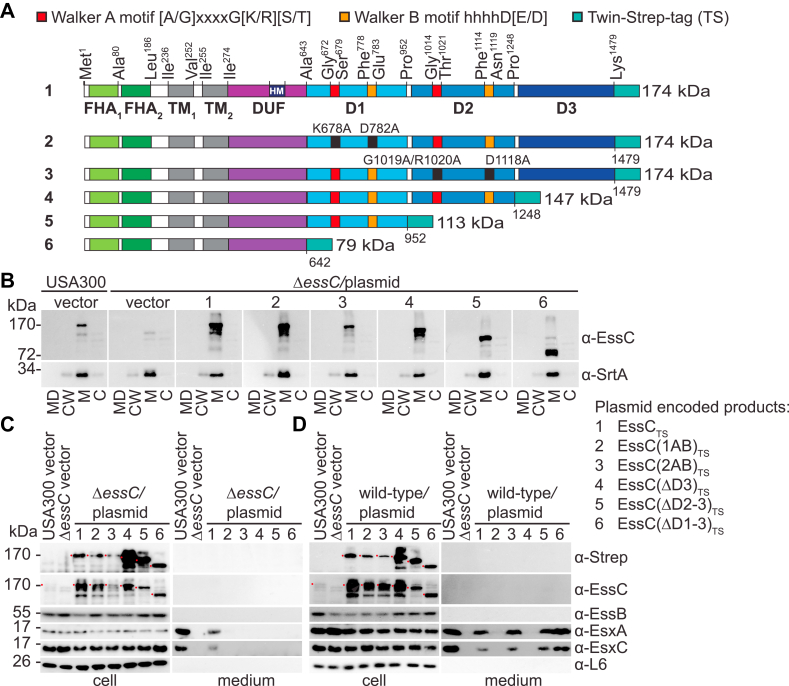
Figure 1.
Purification of the ESS membrane complex of Staphylococcus aureus USA300.A, depiction of domain organization of recombinant EssC and tagged EssCTS proteins. D: ATPase domain (shades of blue); DUF, domain of unknown function (purple); FHA, forkhead-associated domain (light and dark green); TM, transmembrane domain (gray); TS, Twin-Strep tag (turquoise). B, predicted topology of EssC. Colors for each subdomain are as in A. C, cultures of S. aureus fractionated between cells (cell) and culture medium (medium). Proteins in samples were separated by SDS-PAGE and transferred to PVDF membranes for immunoblot analyses, and polyclonal sera are indicated to the right of each blot. Number on top of blot identifies the bacterial culture. Ribosomal protein L6 (α-L6) served as a loading and fractionation control. D and E, DDM extracts were prepared from strains ΔessC pessC (lane 1, control) and ΔessC pessCTS (lane 2) and purified over Strep-Tactin Sepharose resins. Proteins in eluates were separated by SDS-PAGE, and the gel was stained with Coomassie (D) or electrotransferred to PVDF membranes for immunoblot analyses with indicated polyclonal sera (E). Black arrows indicate immunoreactive species corresponding to the expected size of the proteins of interest. Gray arrow indicates an alternative immunoreactive EsaA species. DDM, n-dodecyl β-d-maltoside; ESS, ESAT-6 like secretion system; PVDF, polyvinylidene difluoride.
To gain insights into the biochemical activity of EssC, we sought to purify the protein from membrane fractions of S. aureus. To facilitate purification, the amino-acid sequence WSHPQFEKGGGSGGGSGGSSAWSHPQFEK or Twin-Strep (TS) tag was inserted at the C terminus (Fig. 1A). To verify that the tag did not alter the activity of EssC, a secretion assay was performed (4, 14, 20). USA300 lacking essC (ΔessC) was transformed with plasmids expressing untagged (pessC) and tagged essC (pessCTS). Cultures of the new strains were spun to separate bacterial cells (cell) and the extracellular medium (medium). Cells were lysed with lysostaphin and proteins in cell and medium compartments, separated by SDS-PAGE, and subjected to Western blot analyses (Fig. 1C). The presence of immune reactive species for EsxA and EssD in spent culture media indicated that EssTS complemented the loss of chromosomally encoded EssC. As controls, secretion was abolished in ΔessB and ΔessC strains; since the ribosomal protein L6 (L6) sedimented with cellular fractions exclusively, secretion was not because of nonspecific cell lysis (Fig. 1C). Next, bacterial cultures were used to extract membrane proteins using n-dodecyl β-d-maltoside (DDM), and cleared DDM extracts were applied over Strep-Tactin Sepharose. Bound proteins were eluted with desthiobiotin for LC/MS/MS (Table 1), SDS-PAGE (Fig. 1D), and Western blot (Fig. 1E) analyses. LC/MS/MS revealed an enrichment of peptides for the conserved ESS proteins, EsaA, EssA, EssB, and EssC in ΔessC pessCTS samples as compared with the ΔessC pessC control (Table 1). A few peptides also mapped to EsxA, EsaB, and DUF5079 (encoded by SAUSA300_0295) (Table 1). Owing to their small sizes, EsxA and EsaB digests yielded fewer peptides. DUF5079 is encoded in ESS clusters, but its function is not known; the gene is often duplicated and could encode an orphan immunity factor. A protein encoded by SAUSA300_0274 (four genes upstream of esxA-SAUSA300_0278) was also identified by LC/MS/MS (Table 1). Subsequent analysis of a strain deleted for SAUSA300_0274 demonstrated that the gene is dispensable for T7b secretion (Fig. S2). Further investigation into SAUSA300_0274 was not pursued.
Table 1.
Identification of proteins copurifying with EssC and EssCTS using LC/MS/MS
| Protein | Molecular weight (kDa) | Total number of peptides |
|
|---|---|---|---|
| EssC | EssCTS | ||
| EssC | 170.82 | 43 | 653 |
| EssB | 51.99 | 5 | 108 |
| EsaA | 114.75 | 0 | 47 |
| SAUSA300_0274 | 57.89 | 1 | 29 |
| EssA | 17.38 | 0 | 24 |
| EsaB | 9.11 | 0 | 3 |
| EsxA | 11.03 | 0 | 2 |
| SAUSA300_0295 | 15.64 | 0 | 1 |
SDS-PAGE analysis of Strep-Tactin eluates revealed discrete bands that stained with Coomassie brilliant blue for the EssCTS extract but not the untagged EssC control (Fig. 1D). Similarly, immunoblotting identified the transmembrane proteins EsaA, EssA, and EssB as well as soluble EsxA and EsaB in EssCTS but not from EssC purification, thereby validating the mass spectrometry results (Fig. 1E). Although not identified by LC/MS/MS, the secreted effector EssD, its immunity factor EssI, and putative chaperone EssE were also found in the EssCTS eluate (Fig. 1E). Of note, protease inhibitors were used during the extraction procedure, yet multiple immune reactive species were observed for several proteins, in particular EsaA and EssC (vide infra). It is not clear whether such processing is the result of nonspecific proteolytic activity or regulated proteolysis integral to the T7bSS.
FHA2 recruits EssB and EssA to the translocon
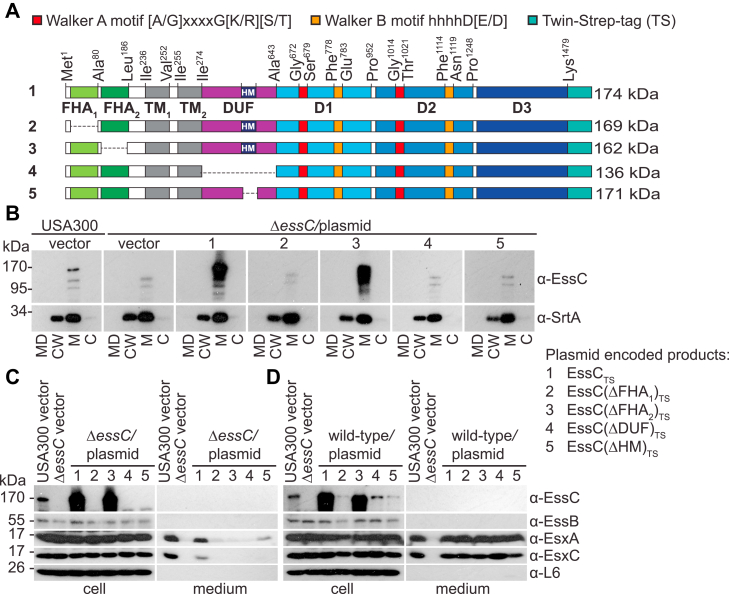
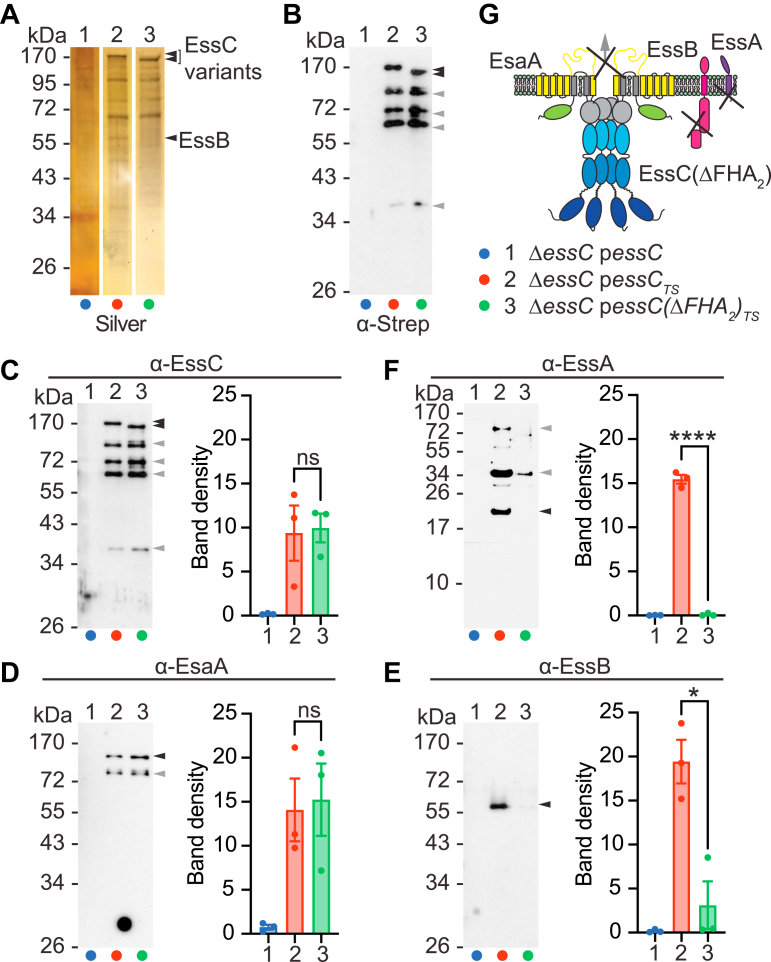
The two FHA domains of EssC share 23.21% identity and are altogether missing in the EccC1–5 of M. tuberculosis as well as in B. anthracis strains that retained EssC but lack EsaA, EssB, EssA, and EsaB (Fig. S1, C and D) (21). Plasmid constructs were generated to produce tagged EssC lacking each one of the FHA domains yielding EssC(ΔFHA1)TS and EssC(ΔFHA2)TS, respectively (Fig. 2A). This analysis was extended to DUF and a small predicted hydrophobic segment (HM) within DUF yielding EssC(ΔDUF)TS and EssC(ΔHM)TS, respectively (Fig. 2A). The four new plasmids were transformed in strain ΔessC for immunoblotting analyses (Fig. 2, B and C). Constructs lacking FHA1, DUF, or HM did not yield any product that could be detected with the anti-EssC polyclonal serum (Fig. 2, B and C). Only EssC(ΔFHA2)TS accumulated to the same extent as EssCTS (Fig. 2, B and C). Subcellular fractionation of bacteria demonstrated that EssC(ΔFHA2)TS retained its membrane location (Fig. 2B) but did not support the secretion of EsxA and EsxC (Fig. 2C). Immunoblotting for the membrane protein Sortase A (SrtA), ribosomal protein L6, and EssB served as controls (Fig. 2, B and C). The four plasmids were also transformed in the wildtype strain. Only EssCTS and EssC(ΔFHA2)TS accumulated to a similar extent (Fig. 2D). Surprisingly, overproduced EssC(ΔFHA2) did not exert a dominant-negative function on EsxA and EsxC secretion (Fig. 2D). Together, the results suggest that the deletions of FHA1, DUF, and HM render EssC unstable. This is in agreement with a report suggesting that deletion of both FHA domains renders EssC unstable (31). This instability can be attributed solely to the loss of FHA1. Stable EssC(ΔFHA2) did not interfere with the ESS translocon, either because it was not recruited to the translocon or because incorporation of these defective subunits was tolerated (Fig. 2D). To look into the role of FHA2 further, DDM solubilized extracts were prepared from cultures of strain ΔessC harboring plasmid-encoded EssC(ΔFHA2)TS as well as EssCTS and EssC for controls. All extracts were purified over Strep-Tactin Sepharose, and proteins in eluates were separated by SDS-PAGE for silver staining (Fig. 3A) and immunoblot analyses (Fig. 3, B–F). As expected, binding to the beads was only observed for tagged proteins (Fig. 3, A–F; lane 1). Both bound EssCTS and EssC(ΔFHA2)TS recruited EsaA (Fig. 3, B–D; lanes 2–3). However, in the absence of FHA2, EssC no longer interacted with EssB and EssA (Fig. 3, E and F; lane 3). Because this partial complex was still embedded in the membrane (Fig. 3G) and cells remained viable, we conclude that incomplete EssC–EsaA translocons are not leaky.
Figure 2.
T7b secretion in mutants lacking FHA domains and DUF.A, illustration of EssCTS and its variants EssC(ΔFHA1)TS, EssC(ΔFHA2)TS, EssC(ΔDUF)TS, and EssC(ΔHM)TS. Color code for each domain is as for Figure 1A. Walker A and B motifs of D1 and D2 are labeled in red and orange, respectively. B, cultures of Staphylococcus aureus strains (as indicated above blot and following numbering code 1–5 to the right) were separated into medium (MD), cell wall (CW), membrane (M), and cytosolic (C) fractions and analyzed by immunoblot using rabbit sera: α-EssC and α-SrtA (as a membrane fractionation control). C and D, cultures of S. aureus, ΔessC (C) or wildtype (USA300, D) bearing the vector control or plasmids (1, 2, 3, 4, 5) as indicated to the right were separated into medium, and cell fractions and proteins in samples were visualized by immunoblot using rabbit sera as indicated to the right of the blots. DUF, domain of unknown function; FHA, forkhead-associated domain.
Figure 3.
Assembly of EssC(ΔFHA2)TSin the ESS membrane complex. DDM extracts were prepared from the indicated strains and purified over Strep-Tactin Sepharose resin. Proteins in eluates were separated by SDS-PAGE, and the gel was stained with silver (A) or electrotransferred to PVDF membranes for immunoblot analyses with α-Strep (B) or with indicated polyclonal sera, α-EssC (C), α-EsaA (D), α-EssB (E), α-EssA (F). Black arrows indicate species for which band densities from three replicate experiments were quantified using ImageJ (54) and analyzed with an unpaired parametric t test. p > 0.1234 (ns), 0.0332 (∗), 0.0021 (∗∗), 0.0002 (∗∗∗), and <0.0001 (∗∗∗∗). Gray arrows indicate alternative immunoreactive species. G, illustration depicting the loss of interactions between EssC(ΔFHA2) and EssB-EssA but not with EsaA. DDM, n-dodecyl β-d-maltoside; ESS, ESAT-6 like secretion system; FHA, forkhead-associated domain; ns, not significant; PVDF, polyvinylidene difluoride.
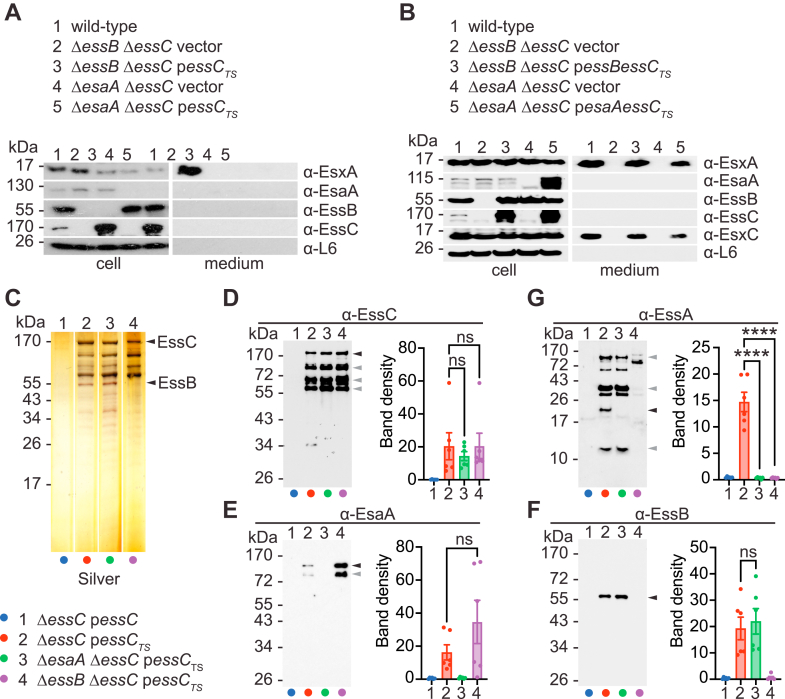
To further explore the formation of partial ESS complexes, the pessCTS plasmid or vector control was introduced in the double mutant strains ΔesaAΔessC and ΔessBΔessC. Cultures of the four new strains were separated into cell and medium fractions and validated by Western blot using sera against EsxA, EsaA, EssB, EssC, as well as secreted EssH and cytosolic ribosomal protein L6 as controls (Fig. 4A). As expected, secretion of EsxA was abrogated in all these strains, but no other differences were noted. For instance, deletion of esaA did not impact the production of EssB and vice versa (Fig. 4A). As additional controls, the double mutant strains ΔesaAΔessC and ΔessBΔessC were complemented with a plasmid expressing either genes esaA and essC (pesaAessCTS) or genes essB and essC (pessBessCTS). In both cases, EsxA and EsxC secretion was restored (Fig. 4B). Next, DDM-soluble fractions of strains bearing pessCTS or vector were purified over Strep-Tactin. Silver staining (Fig. 4C) and Western blotting (Fig. 4D) revealed that comparable amounts of EssCTS were produced in the single ΔessC and the double mutant strains ΔesaAΔessC and ΔessBΔessC (Fig. 4, C and D). As noted earlier, although protease inhibitors were added during sample preparation, four immune reactive species were observed for EssC regardless of the strain background (Fig. 4, C and D). Immunoblot analyses also revealed that EsaA copurified with EssCTS in the absence of EssB (Fig. 4E) and likewise EssB copurified with EssCTS in the absence of EsaA (Fig. 4F). However, EssA no longer associated with EssCTS in the absence of either EsaA or EssB (Fig. 4G). Together, the data suggest that EssC can associate with EsaA and EssB independently, but EssA interaction requires both associations.
Figure 4.
ESS complex assembly in the absence of EsaA or EssB.A and B, cultures of Staphylococcus aureus strains (as indicated on panel) were centrifuged to separate proteins in cell and medium fractions. Proteins in samples were separated by SDS-PAGE and transferred to PVDF membranes for immunoblot analyses, and polyclonal sera are indicated to the right of each blot. C–G, DDM extracts were prepared from strains 1 to 4 with color code listed in B and purified over Strep-Tactin Sepharose resin. Proteins in eluates were separated by SDS-PAGE, and the gel was stained with silver (C) or electrotransferred to PVDF membranes for immunoblot analyses with indicated polyclonal sera α-EssC (D), α-EsaA (E), α-EssB (F), and α-EssA (G). Black arrows indicate species for which band densities from three replicate experiments were quantified using ImageJ (54) and analyzed with an unpaired parametric t test. p > 0.1234 (ns), 0.0332 (∗), 0.0021 (∗∗), 0.0002 (∗∗∗), and <0.0001 (∗∗∗∗). Gray arrows indicate alternative immunoreactive species. DDM, n-dodecyl β-d-maltoside; ESS, ESAT-6 like secretion system; ns, not significant; PVDF, polyvinylidene difluoride.
D2 is required for the assembly of the ESS translocon
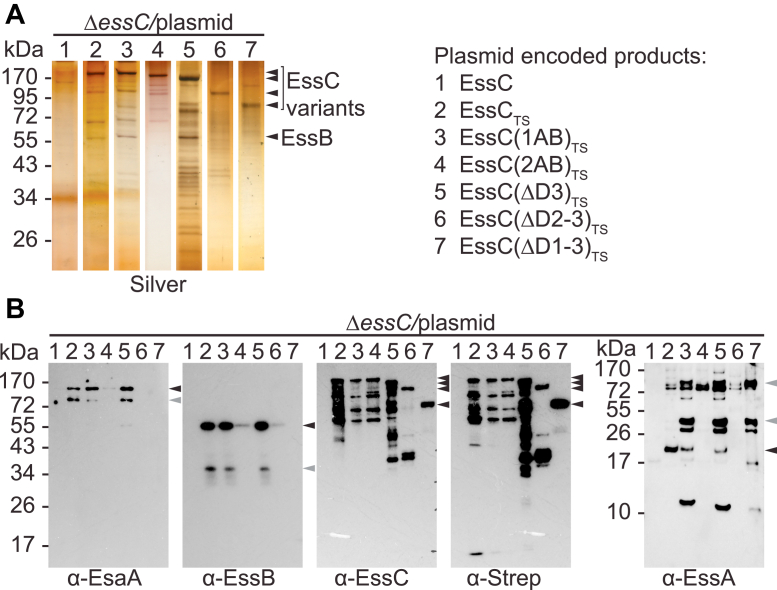
The three FtsK/SpoIIIE-like domains, D1D2D3, complete the C terminus of EssC (Figs. 1B and 5A). Only D1 and D2 contain canonical ATP-binding catalytic Walker A ([A/G]xxxxG[K/R][S/T]) and Walker B (hhhhD[E/D]) motifs. To evaluate the contribution of these domains for T7b secretion, plasmids encoding C-terminally TS-tagged essC variants lacking D3 [EssC(ΔD3)TS], D2 and D3 [EssC(ΔD2–3)TS], or D1 through D3 [EssC(ΔD1–3)TS] were generated (Fig. 5A). Similarly, two variants with substitutions in Walker A and B motifs were also constructed: EssC(1AB)TS with substitutions K678A (Walker A) and D782A (Walker B), and EssC(2AB)TS with substitutions G1019A/R1020A (Walker A) and D1118A (Walker B) (Fig. 5A). Subcellular fractionation of bacteria demonstrated that the new substitutions did not affect the membrane localization of EssC (Fig. 5B). None of the truncated variants or point mutants restored the secretion of EsxA and EsxC in strain ΔessC (Fig. 5C). However, when expressed in the wildtype strain, EssC lacking D3 [EssC(ΔD3)] inhibited the activity of the translocon as documented by the loss of EsxA and EsxC secretion (Fig. 5D). This inhibitory activity was not observed with subsequent deletions of D2 [EssC(ΔD2–3)] and D1 [EssC(ΔD1–3)] (Fig. 5D). Substitutions in Walker A and Walker B motifs of D1, but not D2, also interfered with the activity of the translocon as plasmid expression of essC(1AB)TS resulted in a dominant-negative phenotype over chromosomally expressed essC for the secretion of EsxA and EsxC (Fig. 5D). We speculate that D2 activity is critical for complex formation since variants EssC(1AB) and EssC(ΔD3) (with intact D2) interfered with the wildtype translocon and inhibited T7b secretion, whereas EssC(2AB), EssC(ΔD2–3), and EssC(ΔD1–3) did not. Next, DDM extracts of the ΔessC strain carrying plasmid-encoded EssCTS variants were purified over Strep-Tactin Sepharose and eluates examined by silver staining of SDS-PAGE (Fig. 6A) or immunoblot (Fig. 6B). EssC(ΔD1–3) lacking all three ATPase domains was quite stable as compared with all other variants, including full-length EssC. EssC(ΔD1–3) failed to copurify with EsaA, EssA, and EssB. EssC(ΔD2–3) and EssC(2AB) behaved similarly (Fig. 6B). The absence of D3 or substitutions in the Walker A/B motifs of D1 did not affect the association of EssC with EsaA and EssB (Fig. 6B). In summary, all ATPase domains are required for substrate secretion. However, D2 is also critical for the assembly of the ESS membrane complex.
Figure 5.
T7b secretion in mutants bearing altered ATPase domains.A, domain organization of EssCTS and its variants. Color code for each domain is as indicated for Figure 1A. B, cultures of Staphylococcus aureus strains (as indicated in the figure) were separated into medium (MD), cell wall (CW), membrane (M), and cytosolic (C) fractions and analyzed by immunoblot using rabbit sera: α-EssC and α-SrtA (as a membrane fractionation control). C and D, cultures of S. aureus, ΔessC (C) or wildtype (USA300, D) bearing the vector control or plasmids as indicated to the right were separated into cell and medium fractions, and proteins in samples visualized by immunoblot using rabbit sera as indicated to the right of the blots.
Figure 6.
Purification of ESS complexes with deletions or substitutions in various ATPase domains.A, DDM extracts were prepared from the indicated strains (1, 2, 3, 4, 5, 6, 7) and purified over Strep-Tactin Sepharose resin. Proteins in eluates were separated by SDS-PAGE, and the gel was stained with silver or (B) electrotransferred to PVDF membranes for immunoblot analyses with indicated polyclonal sera. Black arrows indicate immunoreactive species corresponding to the expected size of the proteins of interest. Gray arrows indicate alternative immunoreactive species. DDM, n-dodecyl β-d-maltoside; ESS, ESAT-6 like secretion system; PVDF, polyvinylidene difluoride.
EssC association with EsaA, EssA, and EssB does not require ESS-encoded substrates
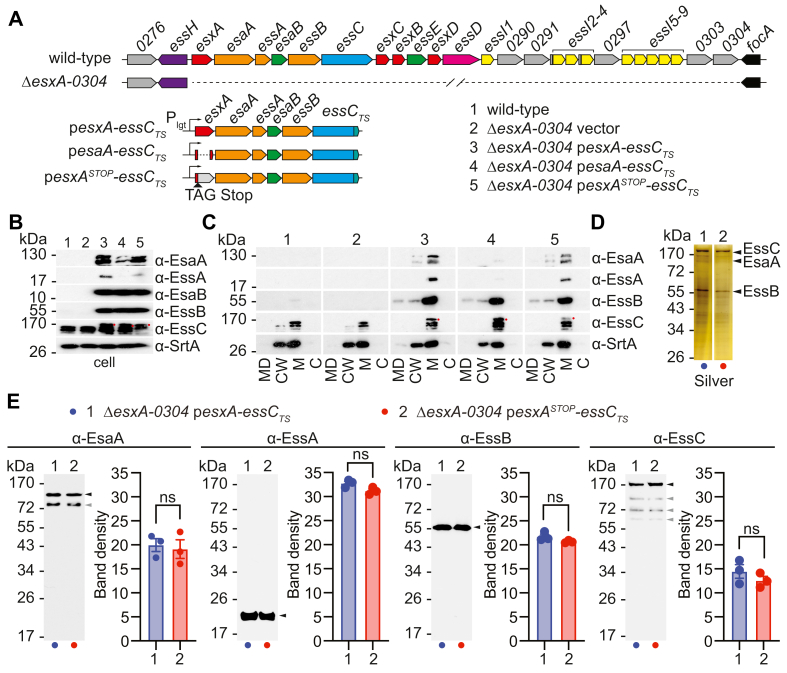
Our results indicated that D2 contributes to the assembly of the ESS translocon. In an independent study, Mietrach et al. (37) used fluorophore-labeled D2D3 and D3 of USA300 EssC (as used in this study) to report an interaction between D2 and purified EsxB. Our LC/MS/MS analysis of purified EssCTS identified the secreted substrate EsxA as a possible interacting partner of the ESS translocon (Table 1). Thus, we wondered if ESS-encoded WXG100 proteins participate in the assembly of the translocon via D2 interactions. To test this possibility, a mutant lacking gene esxA through SAUSA300_0304 (ΔesxA-0304) and thus also lacking esxB, esxC, and esxD, was constructed along with two plasmids, pesxA-essCTS and pesaA-essCTS (Fig. 7A). In this experiment, only one WXG100 protein is provided on plasmid pesxA-essCTS (Fig. 7A). Although expression and production of recombinant genes were driven by the constitutive lgt promoter and a strong Shine–Dalgarno sequence (39), production of EsaA and EssA was not observed when plasmid pesaA-essCTS was introduced in strain ΔesxA-0304 (Fig. 7B). When the clone included gene esxA (plasmid pesxA-essCTS), production of EsaA and EssA was restored (Fig. 7B). When esxA was modified to introduce a stop codon at Glu 10 of EsxA (plasmid pesxASTOP-essCTS), production of EsaA and EssA in strain ΔesxA-0304 was also restored (Fig. 7, A and B). Subcellular fractionation experiments confirmed that plasmid-borne EsaA, EssA, EssB, and EssC retained their membrane localization in strain ΔesxA-0304 (Fig. 7C). Next, TS-tagged proteins were purified from cultures of strain ΔesxA-0304 carrying plasmid pesxA-essCTS or pesxASTOP-essCTS (Fig. 7D). EsaA, EssA, and EssB copurified with EssCTS (Fig. 7, D and E). The data show that ESS-encoded WXG and LXG substrates are not required for the assembly of the T7b membrane complex. Nonetheless, in S. aureus, USA300 esxA bears sequence information that is important for the optimal expression of downstream genes esaA and essA. The lost information cannot be compensated by providing a strong constitutive promoter and Shine–Dalgarno sequence.
Figure 7.
Purification of the ESS membrane complex in the absence of secreted substrates.A, illustration of strains and plasmids used in experiments shown in the figure. B, Western blot analyses of cell extracts from strains (1, 2, 3, 4, 5) depicted in A. C, cultures of Staphylococcus aureus strains (1, 2, 3, 4, 5) were separated into medium (MD), cell wall (CW), membrane (M), and cytosolic (C) fractions and analyzed by immunoblot using rabbit sera as indicated to the right of the blots. D, DDM extracts were prepared from two strains as indicated above the panel and purified over Strep-Tactin Sepharose resin. Proteins in eluates were separated by SDS-PAGE, and the gel was stained with silver or (E) electrotransferred to PVDF membranes for immunoblot analyses with indicated polyclonal sera. Black arrows indicate species for which band densities from three replicate experiments were quantified using ImageJ (54) and analyzed with an unpaired parametric t test. p > 0.1234 (ns). Gray arrows indicate alternative immunoreactive species. DDM, n-dodecyl β-d-maltoside; ESS, ESAT-6 like secretion system; ns, not significant; PVDF, polyvinylidene difluoride.
Discussion
The T7a and T7b secretion systems borrow an ATPase from the FtsK/SpoIIIE family of the FtsK–HerA superfamily of P-loop ATPases within the larger AAA+ superfamily (ATPases Associated with various cellular Activities) (40, 41, 42). Characteristics of the AAA+ superfamily include the presence of a phosphate-binding loop (P-loop) or Walker A motif ([A/G]xxxxG[K/R][S/T]) forming the nucleotide-binding pocket and Walker B motif (hhhhD[E/D]) and arginine finger (R-finger) for ATP hydrolysis (41, 43). ATPase activity requires interaction between adjacent subunits forming a ring with Walker residues contributed by the cis subunit and R-finger contributed by the adjacent trans subunit (43). Thus, oligomerization completes the ATP-binding pocket, with most AAA+ proteins forming ring-shaped hexamers for ATP hydrolysis–driven translocation of protein or DNA substrates through a central channel. Protein translocation by AAA+ ATPases results in unfolding, whereas DNA translocation leads to unwinding during replication or packaging of viral genomes (40, 41, 42, 44). For example, FtsK proteins assemble into membrane-tethered ring-shaped hexamers at bacterial septa to translocate naked double-stranded DNA into daughter cells. FtsK activity can be described as treadmilling along the spiral staircase DNA substrate, a process essentially like cytomotive protein filaments, but in a closed circular arrangement (45, 46).
As far as we know, EssC proteins of Firmicutes translocate proteins, not DNA. Like the related EccC (T7a) of Actinomyces and unlike cell division FtsK proteins, T7b EssC proteins contain multiple structural ATP-binding domains. By tagging EssC and using a combination of mass spectroscopic and immunoblot analyses, we determined that EssC copurifies with EsxA, EsaA, EssA, EsaB, and EssB following detergent extraction of membrane proteins from wildtype S. aureus. By generating a strain unable to produce EsxA and downstream WXG/LXG products, we ruled out the possibility that these secreted substrates are required for the assembly of this DDM extractable complex. Using similar approaches, we asked how each subdomain may contribute to secretion and complex assembly. We generated EssC variants lacking either FHA1, FHA2, DUF or with increasing C-terminal deletions (ΔD3, ΔD2–3, and ΔD1–3) or with mutations in Walker A/B motifs of D1 or D2. None of these variants supported protein secretion as reported by others (31, 37). Removal of FHA1 or DUF, including the small HM segment within DUF, failed to produce EssC altogether. We do not have an explanation for these observations. We found that EssC associates with EssB and EssA via FHA2, a unique structural feature of T7bSS (Fig. 8, A and B). FHA domains are commonly found in eukaryotes where they often participate in signal transduction processes and interact with phosphorylated ligands to modulate cell cycle, chromosome segregation, and protein degradation (47). Only a few FHA-containing bacterial proteins have been characterized thus far. In the plant pathogen Agrobacterium tumefaciens, the protein known as Fha binds phosphothreonine–modified TssL, an inner-membrane core component of the type 6 secretion system, affecting its assembly and secretion (48). Here, we found that EssC lacking FHA2 does not associate with EssB and EssA but retains interactions with EsaA (Fig. 8B). While we are not yet aware of phosphothreonine or phosphoserine modification in T7b proteins, the cytosolic domain of EssB has been shown to adopt a pseudokinase fold and mediate dimerization of EssB (49). Studies in Bacillus subtilis have shown that the isolated pseudokinase domain of YukC (EssB homolog) and FHA1–2 of YukB (EssC homolog) interact (50). Thus, in S. aureus, the pseudokinase and FHA2 domains might also be sufficient to establish an interaction between EssB and EssC to prompt EssA recruitment. Indeed, EssA did not associate with the translocon in the absence of EssB.
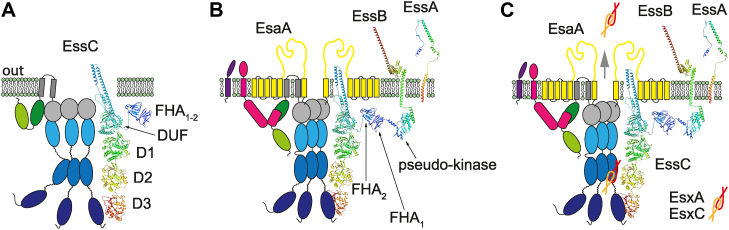
Figure 8.
Modeling the ESS membrane complex.A, EssC multimerization requires a functional D2 domain (shade of blue). B, EssC interacts via its FHA2 domain (shade of green) with the cytosolic domain of EssB (pink) encompassing a pseudokinase fold. The exact association between EssC, EsaA, and EssA remains to be determined. Assembly between EssC, EsaA, EssA, and EssB does not require ESS-encoded WXG100 substrates but is lost when the Walker A and B motifs of D2 are mutated. C, substrates interact with the ESS membrane complex, which directs them for secretion. Protein structures for EssC, EssB, and EssA were predicted using AlphaFold (55). ESAT-6 like secretion system; FHA, forkhead-associated domain.
When focusing on the C terminus of EssC, we found that D3 is dispensable for complex assembly since the truncated variant, EssC(ΔD3), pulled down the EsaA–EssA–EssB complex. EssC(ΔD3) also exerted a dominant-negative phenotype for secretion likely because it was able to form mixed EssC(ΔD3)–EssC–EsaA–EssA–EssB complexes. When we examined other mutants, EssC(ΔD2–3), EssC(ΔD1–3), or substitutions in the Walker A/B motifs of D1 or D2, we found that the dominant-negative phenotype correlated with the ability to pull down EsaA–EssA–EssB. Our results suggest that the Walker A/B motifs of D2 are critical for bringing or keeping the ESS membrane complex (EssC–EsaA–EssA–EssB) together. While not described here, we failed to measure ATP hydrolysis using recombinant full-length EssC or smaller fragments regardless of incubation with WXG100 substrates. Zolner et al. (31) solved the structure of D2D3 of G. thermodenitrificans GtEssC (GtD2D3) and found intact ATP in the nucleotide-binding site of GtD2 but not GtD3, which points to weak intrinsic ATPase activity. Mietrach et al. (37) reported the crystal structures of S. aureus D3 (same SaD3 as the one used in this study) and by comparing structures of GtEssC and T. curvata EccC (TcEccC) proposed that the ATP-binding sites for SaD3 and GtD3 may be structurally occluded as compared with T7a D3 domains (35, 37). D3 domains in all three species (Gt, Sa, and Tc) carry a degenerated Walker B motif with no distinguishable R finger. D3 of S. aureus could be mutated to either increase or reduce T7b secretion once again suggesting a contribution for ATP binding or hydrolysis (37). In T. curvata, the soluble TcD1D2D3 domain was shown to form dimers and higher-order oligomers in the presence of TcEsxB, whereas TcEsxA resulted in oligomer dissociation in vitro (35). A model was proposed whereby substrates with a C-terminal secretion motif (e.g., EsxB of M. tuberculosis or T. curvata) dock into the exposed nucleotide-binding site of TcD3, triggering a conformational change transduced along the EccC monomer (35). In this model, ATPases are inhibited by subunit interactions along the EccC monomer, and the substrate relieves inhibition as it is moved upward. This model is different from the proposed mechanism of canonical FtsK (AAA+ ATPases) whereby conformational changes are transduced via the R-finger when subunits are brought together in the hexameric ring.
Secretion motifs in staphylococcal T7b substrates have been difficult to pinpoint (14), yet Warner et al. (51) noted that highly variable sequences were associated with D3 and immediate downstream genes encoding strain-specific WXG and LXG effectors. Swapping of D3 between strains supported the notion of EssC selectivity toward nonconserved effectors but not toward conserved EsxA (36). Thus, D3 is likely to play a role in substrate recruitment. Yet, isolated D2 has also been shown to interact with EsxB and EsxC of S. aureus (37). We surmise that oligomerization of SaEssC may first be imposed by its 12 membrane-spanning segments. This is modeled on the proposed structures of EccC3 and EccC5 revealed by recent cryo-EM images of mycobacterial ESX-3 and ESX-5 complexes (34, 52). In S. aureus, at least two EsaA molecules (each containing seven transmembrane segments) could be wrapped around the EssC hexamer (Fig. 8C). Interestingly, DUF may also adopt a Rossman fold as demonstrated for EccC3 of M. smegmatis ESX-3 (34). Thus, the C terminus of SaEssC may contain four domains with similar structures (D0–D1–D2–D3), with D1 and D2 carrying canonical Walker A and B motifs and R-fingers. We propose that R734 and R1078 in D1 and D2 of SaEssC allow for the formation of two stacked rings under the transmembrane and D0 segments, introducing rigidity into EssC hexamers and perhaps providing for opening and closing into a conduit that extends across the plasma membrane (Fig. 8C). The Walker A/B motifs of D2 are essential for oligomerization and assembly of the ESS complex, and D0 is required for protein stability. How other proteins in this complex, ATP and ultimately secreted substrates, influence conformational changes and gating of this complex remain to be elucidated.
Experimental procedures
Media and growth conditions
S. aureus strains were cultured in tryptic soy broth or tryptic soy agar at 37 °C, unless otherwise stated, and media were supplemented with 10 μg/ml chloramphenicol for plasmid selection and 0.2% heat-inactivated horse serum (Gibco/Life Technologies) to trigger secretion.
Bacterial strains and plasmids
Relevant strains and plasmids used in this study are listed in Table S1 and primers in Table S2. S. aureus USA300 LAC∗, a clone of the epidemic community–acquired methicillin-resistant S. aureus strain (38) was used as the wildtype strain. Plasmid DNA was passaged in S. aureus RN4220 before transformation into USA300 strains. Mutations in strain USA300 LAC∗ were introduced by pKOR1-mediated allelic replacement (53). Plasmids pKOR1-ΔessC, pKOR1-ΔessBΔessC, and pKOR1-ΔesxA-0304 were constructed by PCR amplifying upstream and downstream DNA sequences flanking the desired region to be deleted. The following primer pairs were used for amplification: pKOR1-ΔessC primers 112F/112R and 128F/128R; pKOR1-ΔessBΔessC primers 106F/106R and 107F/107R; and pKOR1-ΔesxA-0304 primers 97F/97R-XbaI and 98F-XbaI/98R. DNA fragments for pKOR1-ΔessC and pKOR1-ΔessBΔessC were stitched by PCR and for pKOR1-ΔesxA-0304 by restriction digestion using XbaI and ligation. Stitched DNA fragments were cloned into pKOR1 using BP Clonase Kit (Invitrogen). Mutants ΔessC, ΔessBΔessC, and ΔesxA-0304 were constructed as described (53). Mutant ΔesaAΔessC was generated by transforming ΔesaA strain with pKOR1-ΔessC, and ΔessC was introduced by allelic replacement. Plasmids pessC and pessCTS were constructed by PCR amplification of essC gene from S. aureus USA300 LAC∗ genomic DNA using primer pairs 557/572 or 557/563, respectively. PCR products were cloned into the pWWW412 vector at the XhoI/BamHI sites (39). Plasmids pessC(ΔD3)TS, pessC(ΔD2-3)TS, and pessC(ΔD1-3)TS were constructed using pessCTS as a template for PCR amplification of intermediate linear products using primers pairs 661/662, 660/662, and 659/662, respectively. Linear PCR products were digested with BamHI (as well as DpnI) and religated to generate circular plasmids encoding truncated essCTS variants. Plasmid pessC(1AB)TS was constructed by PCR amplification and Gibson assembly (NEB) of four DNA fragments from pessCTS template using four primer pairs 663/670, 665/666, 667/668, and 669/664. Plasmid pessC(2AB)TS was constructed by QuikChange PCR mutagenesis using pessCTS as template. Primer pair 156Fmut/156Rmut was used to introduce G1019A,R1020A in Walker A and 157Fmut/157Rmut for D1118A in Walker B. Plasmids pessC(ΔFHA1)TS, pessC(ΔFHA2)TS, pessC(ΔDUF)TS, and pessC(ΔHM)TS were constructed using pessCTS as a template for PCR amplification of intermediate linear product using overlapping 5′-phosphorylated primer pairs 173F-P/173R-P, 174F-P/174R-P, 175F-P/175R-P, and 176F-P/176R-P, respectively. Linear PCR products were religated to generate circular plasmids encoding truncated essCTS variants. Plasmids pesxA-essCTS, pesaA-essCTS, and pesxASTOP-essCTS were constructed by PCR amplification of esxA-essC, esaA-essC, and esxASTOP-essC fragments from S. aureus USA300 LAC∗ genomic DNA using primer pairs 121F/121R, 135F/121R, and 203F/121R, respectively. XhoI/BglII-digested DNA products and XhoI/BamHI-digested pessCTS plasmid were ligated using DNA Ligase (NEB) to generate pesxA-essCTS, pesaA-essCTS, and pesxASTOP-essCTS. To generate plasmids pesaAessCTS and pessBessCTS (these two plasmids encode two genes each), plasmid pesxASTOP-essCTS was used as a template, and linear products were obtained by PCR using overlapping 5′-phosphorylated primer pairs 230F-P/230R-P and 231F-P/231R-P. The primer pairs were designed to eliminate the intervening essA-esaB-essB and esaA-essA-esaB encoding sequences, respectively.
Fractionation of bacterial cultures
For protein secretion and subcellular fractionation assays, S. aureus culture aliquots were spun at 10,000g for 10 min to separate proteins in the supernatant (medium) and cells. To assess for protein secretion, cells in bacterial pellets were washed and lysed using lysostaphin (10 μg/ml for 1 h at 37 °C) yielding lysed cell extracts. Proteins in these suspensions (medium or cell) were precipitated by addition of 12% final concentration trichloroacetic acid, washed in cold acetone, and dried. For subcellular localization of proteins, washed cells were suspended in 50 mM Tris–HCl, pH 7.5, 0.5 M sucrose, and 10 mM MgCl2 buffer prior to lysostaphin treatment. Samples were spun at 15,000g for 10 min, the supernatants (cell wall fraction) were transferred to a new tube, and protoplasts were lysed by repeated freeze thawing (three cycles). The extracts were spun at 100,000g for 40 min at 4 °C to separate the supernatants containing cytosolic proteins (cytoplasmic fraction). Pellets containing membrane proteins were suspended in PBS buffer (membrane fraction). Proteins in all fractions were precipitated with trichloroacetic acid, washed with methanol, and dried. All precipitates were solubilized in 100 μl of 0.5 M Tris–HCl (pH 8.0), 4% SDS, and heated at 90 °C for 10 min.
Purification of the ESS membrane complex
Bacterial cultures were grown to an absorbance of ∼3.0 at 600 nm at 37 °C. Cells were collected by centrifugation and suspended in buffer A (20 mM Tris [pH 8.0], 300 mM NaCl, 10% [v/v] glycerol, SIGMAFAST Protease Inhibitors [SIGMA]). Cells were lysed using bead beating, and debris was removed by centrifugation. Soluble and insoluble fractions were separated by centrifugation at 100,000g for 1 h. Pellet fractions containing membrane proteins were suspended in buffer A with 0.25% DDM and incubated for 1 h at 4 °C. Insoluble material was removed by centrifugation at 100,000g for 30 min, and cleared supernatants were loaded onto 1 ml bed of Strep-Tactin Sepharose resin (IBA). The resin was washed with 10 ml buffer A and incubated for 18 h with 10 ml of buffer A containing 0.5 g/ml Amphipol A8-35 (Anatrace) to replace DDM. The resin was washed with an additional 10 ml of buffer A containing 0.5 g/ml Amphipol A8-35 prior to elution of proteins with buffer A containing 0.5 g/ml Amphipol A8-35 and 5 mM d-desthiobiotin (IBA). Aliquots of eluted fractions were mixed with an equal volume of sample buffer and separated on 12 or 15% SDS-PAGE and stained with Coomassie and silver or transferred to polyvinylidene difluoride membrane for immunoblot analyses.
Western blot
Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membrane. The membranes were blocked for 1 h at room temperature in 10 ml of blocking buffer containing 0.5% fat-free milk prior to the addition of primary polyclonal antibodies (at a dilution of 1:2000). Membranes were incubated for an additional hour, washed three times for 10 min in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween-20, and incubated with 1:10,000 anti-rabbit or antimouse horseradish peroxidase–conjugated secondary antibody (Cell Signaling Technology) for 1 h. Membranes were washed again in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween-20, and immunoreactive products were revealed by chemiluminescent detection using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Blots were developed on Amersham Hyperfilm ECL (GE Healthcare Life Sciences), and band densities were quantified using ImageJ (National Institutes of Health) (54) software. Graphing and statistical analysis were performed using Prism 9 software (GraphPad Software, Inc).
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information (20, 25, 38, 53, 56, 57).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank members of our laboratory for insightful comments and Chloe Schneewind for careful editing of the article. We remember Olaf Schneewind for his insights and unbounded enthusiasm for all things relating to secretion systems. Work on protein secretion in our laboratory is supported by AI038897 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Infectious Disease Branch.
Author contributions
M. B., S. Y. O., and D. M. conceptualization; M. B., S. Y. O., and D. M. methodology; M. B. and D. M. formal analysis; M. B. and S. Y. O. investigation; M. B. and S. Y. O. data curation; M. B. and D. M. writing–original draft.
Funding and additional information
M. B. was supported by the National Institute of Allergy and Infectious Diseases, United States, National Institutes of Health, United States, postdoctoral fellowship award (F32AI085709). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karen Fleming
Supporting information
References
- 1.Lee V.T., Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15:1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- 2.Green E.R., Mecsas J. Bacterial secretion systems: an overview. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Troman L., Collinson I. Pushing the envelope: the mysterious journey through the bacterial secretory machinery, and beyond. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.782900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burts M.L., Williams W.A., DeBord K., Missiakas D.M. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pallen M.J. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 6.Poulsen C., Panjikar S., Holton S.J., Wilmanns M., Song Y.H. WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gey Van Pittius N.C., Gamieldien J., Hide W., Brown G.D., Siezen R.J., Beyers A.D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdallah A.M., Gey van Pittius N.C., Champion P.A., Cox J., Luirink J., Vandenbroucke-Grauls C.M., et al. Type VII secretion--mycobacteria show the way. Nat. Rev. Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 9.Schneewind O., Missiakas D.M. Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman L., Palmer T. The type VII secretion system of Staphylococcus. Annu. Rev. Microbiol. 2021;75:471–494. doi: 10.1146/annurev-micro-012721-123600. [DOI] [PubMed] [Google Scholar]
- 11.Tran H.R., Grebenc D.W., Klein T.A., Whitney J.C. Bacterial type VII secretion: an important player in host-microbe and microbe-microbe interactions. Mol. Microbiol. 2021;115:478–489. doi: 10.1111/mmi.14680. [DOI] [PubMed] [Google Scholar]
- 12.Ates L.S., Houben E.N.G., Bitter W. Type VII secretion: a highly versatile secretion system. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0011-2015. [DOI] [PubMed] [Google Scholar]
- 13.Bottai D., Groschel M.I., Brosch R. Type VII secretion systems in gram-positive bacteria. Curr. Top Microbiol. Immunol. 2017;404:235–265. doi: 10.1007/82_2015_5015. [DOI] [PubMed] [Google Scholar]
- 14.Anderson M., Aly K.A., Chen Y.H., Missiakas D. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol. Microbiol. 2013;90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundaramoorthy R., Fyfe P.K., Hunter W.N. Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J. Mol. Biol. 2008;383:603–614. doi: 10.1016/j.jmb.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abd El-Fatah R.M., Mesbah N.M., Abo-Elmatty D.M., Aly K.A. The C-terminus of the ESAT6-like secretion system virulence factor EsxC mediates divalent cation-dependent homodimerization. Toxicon. 2018;146:129–137. doi: 10.1016/j.toxicon.2018.02.047. [DOI] [PubMed] [Google Scholar]
- 17.Ilghari D., Lightbody K.L., Veverka V., Waters L.C., Muskett F.W., Renshaw P.S., et al. Solution structure of the Mycobacterium tuberculosis EsxG.EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes. J. Biol. Chem. 2011;286:29993–30002. doi: 10.1074/jbc.M111.248732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renshaw P.S., Lightbody K.L., Veverka V., Muskett F.W., Kelly G., Frenkiel T.A., et al. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 2005;24:2491–2498. doi: 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y., Tan K., Chhor G., Butler E.K., Jedrzejczak R.P., Missiakas D., et al. EsxB, a secreted protein from Bacillus anthracis forms two distinct helical bundles. Protein Sci. 2015;24:1389–1400. doi: 10.1002/pro.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burts M.L., DeDent A.C., Missiakas D.M. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garufi G., Butler E., Missiakas D. ESAT-6-like protein secretion in Bacillus anthracis. J. Bacteriol. 2008;190:7004–7011. doi: 10.1128/JB.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., Iyer L.M., Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucl. Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhe Z.C., Low D.A., Hayes C.S. Polymorphic toxins and their immunity proteins: diversity, evolution, and mechanisms of delivery. Annu. Rev. Microbiol. 2020;74:497–520. doi: 10.1146/annurev-micro-020518-115638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson M., Chen Y.H., Butler E.K., Missiakas D.M. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J. Bacteriol. 2011;193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson M., Ohr R.J., Aly K.A., Nocadello S., Kim H.K., Schneewind C.E., et al. EssE promotes Staphylococcus aureus ESS-dependent protein secretion to modify host immune responses during infection. J. Bacteriol. 2017;199 doi: 10.1128/JB.00527-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohr R.J., Anderson M., Shi M., Schneewind O., Missiakas D. EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. J. Bacteriol. 2017;199 doi: 10.1128/JB.00528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Z., Casabona M.G., Kneuper H., Chalmers J.D., Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2016;2 doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulhuq F.R., Gomes M.C., Duggan G.M., Guo M., Mendonca C., Buchanan G., et al. A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc. Natl. Acad. Sci. U. S. A. 2020;117:20836–20847. doi: 10.1073/pnas.2006110117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aly K.A., Anderson M., Ohr R.J., Missiakas D. Isolation of a membrane protein complex for type VII secretion in Staphylococcus aureus. J. Bacteriol. 2017;199 doi: 10.1128/JB.00482-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoltner M., Ng W.M., Money J.J., Fyfe P.K., Kneuper H., Palmer T., et al. EssC: Domain structures inform on the elusive translocation channel in the type VII secretion system. Biochem. J. 2016;473:1941–1952. doi: 10.1042/BCJ20160257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y., Kuroda M., Yasutake Y., Yao M., Tsumoto K., Watanabe N., et al. Crystal structure analysis reveals a novel forkhead-associated domain of ESAT-6 secretion system C protein in Staphylococcus aureus. Proteins. 2007;69:659–664. doi: 10.1002/prot.21302. [DOI] [PubMed] [Google Scholar]
- 33.Famelis N., Rivera-Calzada A., Degliesposti G., Wingender M., Mietrach N., Skehel J.M., et al. Architecture of the mycobacterial type VII secretion system. Nature. 2019;576:321–325. doi: 10.1038/s41586-019-1633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poweleit N., Czudnochowski N., Nakagawa R., Trinidad D.D., Murphy K.C., Sassetti C.M., et al. The structure of the endogenous ESX-3 secretion system. Elife. 2019;8 doi: 10.7554/eLife.52983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg O.S., Dovala D., Li X., Connolly L., Bendebury A., Finer-Moore J., et al. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jager F., Kneuper H., Palmer T. EssC is a specificity determinant for Staphylococcus aureus type VII secretion. Microbiology (Reading) 2018;164:816–820. doi: 10.1099/mic.0.000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mietrach N., Damian-Aparicio D., Mielich-Suss B., Lopez D., Geibel S. Substrate interaction with the EssC coupling protein of the type VIIb secretion system. J. Bacteriol. 2020;202 doi: 10.1128/JB.00646-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diep B.A., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 39.Bubeck Wardenburg J., Williams W.A., Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucl. Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 43.Wendler P., Ciniawsky S., Kock M., Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 44.Jessop M., Felix J., Gutsche I. AAA+ ATPases: Structural insertions under the magnifying glass. Curr. Opin. Struct. Biol. 2021;66:119–128. doi: 10.1016/j.sbi.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jean N.L., Rutherford T.J., Lowe J. FtsK in motion reveals its mechanism for double-stranded DNA translocation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:14202–14208. doi: 10.1073/pnas.2001324117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massey T.H., Mercogliano C.P., Yates J., Sherratt D.J., Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Durocher D., Jackson S.P. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 48.Lin J.S., Wu H.H., Hsu P.H., Ma L.S., Pang Y.Y., Tsai M.D., et al. Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoltner M., Fyfe P.K., Palmer T., Hunter W.N. Characterization of Staphylococcus aureus EssB, an integral membrane component of the type VII secretion system: atomic resolution crystal structure of the cytoplasmic segment. Biochem. J. 2013;449:469–477. doi: 10.1042/BJ20121209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tassinari M.D.,T., Bellinzoni M., Chabalier M., Ben-Assaya M., Martinez M., Gaday Q., et al. Central role and structure of the membrane pseudokinase YukC in the antibacterial Bacillus subtilis Type VIIb Secretion System. bioRxiv. 2020 doi: 10.1101/2020.1105.1109.085852. [preprint] [DOI] [Google Scholar]
- 51.Warne B., Harkins C.P., Harris S.R., Vatsiou A., Stanley-Wall N., Parkhill J., et al. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17:222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunduc C.M., Fahrenkamp D., Wald J., Ummels R., Bitter W., Houben E.N.G., et al. Structure and dynamics of a mycobacterial type VII secretion system. Nature. 2021;593:445–448. doi: 10.1038/s41586-021-03517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae T., Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Met. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreiswirth B.N., Lofdahl S., Betley M.J., O’Reilly M., Schlievert P.M., Bergdoll M.S., et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 57.Schneewind O., Model P., Fischetti V.A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.