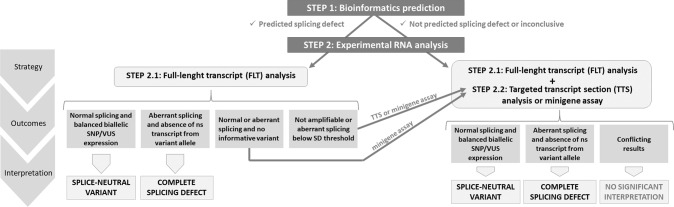
Fig. 1. Flowchart of consecutive splicing analyses recommended by the EMMR-WG to investigate MMR variants.
Bioinformatic predictions are included as a first step because not predicted splicing aberrations must be confirmed in an additional RNA assay, according to InSiGHT MMR variant classification guidelines (v2.4). The full-length transcript (FLT) analysis is suggested as the second step, with the exception that no patient material is available, or the cDNA is not amplifiable as FLT. In these cases, a target transcript section (TTS) analysis or a minigene assay should be performed in the next step. According to InSiGHT MMR variant classification rules, normal splicing and balanced biallelic expression of a variant in the FLT analysis assign variants as splice-neutral. In absence of an informative variant, the effect of intronic variants on RNA splicing has to be further investigated in a minigene assay. This also applies for inconclusive results from FLT analyses. In contrast, if the FLT analysis demonstrates aberrant splicing and excludes the generation of full-length transcript derived from the variant allele, then the variant can be assigned as complete splice defect, especially if bioinformatics also predicted a splicing defect. Otherwise, its presence has to be verified with an independent method, which can be a TTS analysis in another laboratory, or a minigene assay. Additional analyses are also needed when FLT analyses yield inconclusive results.