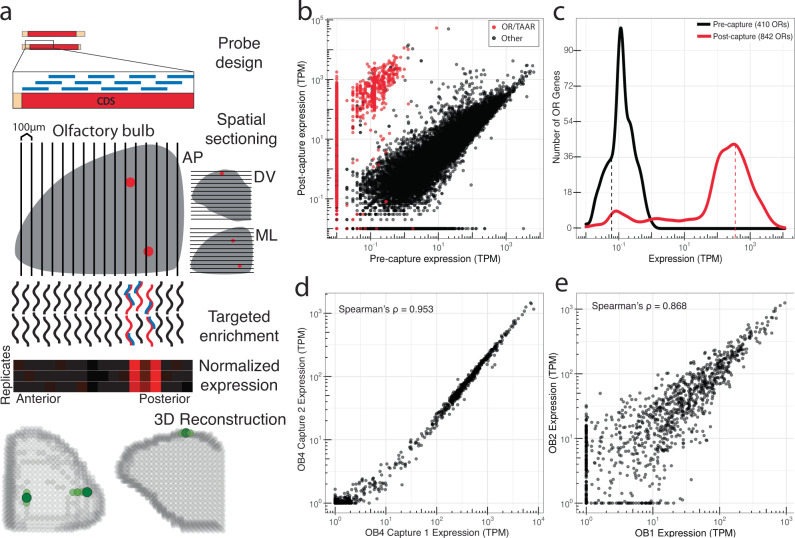
Fig. 1. Target capture sequencing consistently enriches OR transcripts.
a Methodological overview for targeted enrichment of OR and TAAR transcripts from OB sections. Briefly, RNA is extracted from OB tissue and used for cDNA synthesis and library preparation. Biotinylated capture probes are designed against coding sequences (CDS) of interest, enabling enrichment of target genes following streptavidin binding and washing steps. RNA-Seq of enriched libraries allows for high-throughput positional analyses when combined with systematic tissue sectioning in both individual dimensions and in 3D after reconstruction. b Pre- and post-capture normalized abundances (transcripts per million; TPM) of intact OR and TAAR genes (red) and other intact genes (black) from a whole OB. Source data provided as a Source Data file. c Frequency distribution of OR gene abundances pre- and post-capture from a whole OB. d Technical replicates demonstrating OR and TAAR gene abundances from independent capture enrichments of the same whole-OB RNA. e Biological replicates demonstrating OR and TAAR gene abundances from capture enrichment of whole-OB RNA samples from different animals.