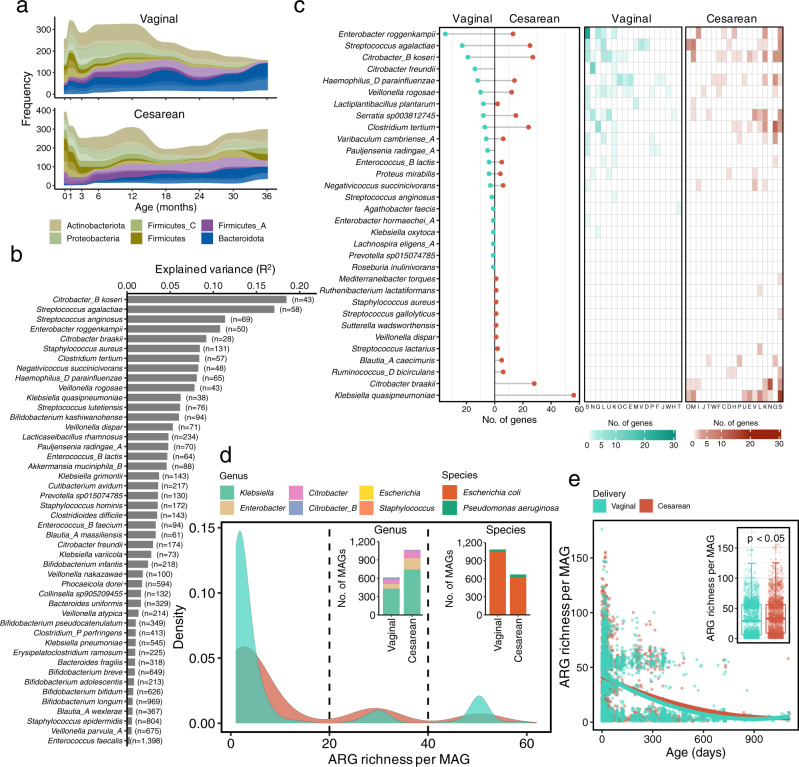
Fig. 5. Influences of delivery mode on early-life gut microbiome at a genome-resolved level.
a Prevalence of 16 bacterial genera in children stratified by delivery mode over time, where each genus was colored by its phylum. Only genera with >10% prevalence in children born by any of delivery modes are plotted. b The explained variance (R2) contributed by delivery mode of 46 species that were significantly (PERMANOVA, FDR < 0.05) associated with delivery mode based on the hamming distance of core genes per species. The number in parentheses indicate the number of MAGs of this species. c The number of genes that were prevalent in C-section born children or vaginally born children (>70% in C-section born children and <30% in vaginally born children, and vice versa) for each species and their associated functions annotated by COGs database. d The density of antibiotic resistance genes (ARGs) richness in each genome of ELGG, and the taxonomic assignment of the genomes at genus (left inset) and species (right inset) level. e The dynamics of ARGs richness from the early-life human gut microbiome in response to the age of children. The gut microbiome from children born by C-section carried higher (two-tailed Wilcoxon test, p < 0.05, inset) number of ARGs than that of children born vaginally. The inset boxes show the interquartile range (IQR), with the horizonal line as the median, the whiskers indicating the range of the data (up to 1.5× IQR), and points beyond the whiskers as outliers.