Figure 4.
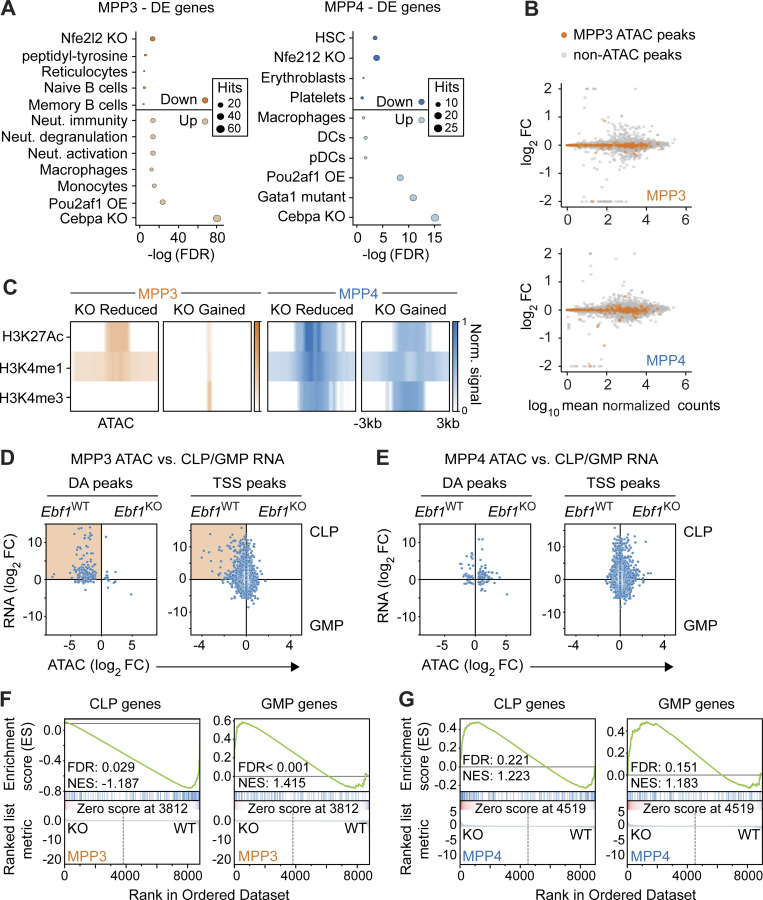
EBF1-dependent chromatin sites in MPP3 cells are associated with expression in CLPs. (A) Selected terms are represented from enrichment analysis of DE genes (FDR < 0.1) between Ebf1WT and Ebf1KO MPP3 (left) and MPP4 (right) cells. Enrichr datasets (GO_Biological_Process_2021, PanglaoDB_Augmented_2021, Gene_Perturbations_from_GEO_down) were used for enrichment analysis. Biological replicates n = 4. (B) Expression analysis of genes annotated to KO Reduced MPP3 peaks identified in ATAC analysis, in Ebf1WT and Ebf1KO MPP3 (upper panel), and MPP4 (lower panel) cells. The x axis represents log10 mean normalized counts of all genes. The y axis represents shrunken |log2 fold change| values of all genes in Ebf1WT versus Ebf1KO MPP3 (upper panel), and MPP4 (lower panel) cells, and is capped at 2. Genes in orange indicate the genes annotated to EBF1-dependent accessibility sites in MPP3 cells. (C) Heatmap displaying average ChIP signal for chromatin marks at DA peaks. Original data from Lara-Astiaso et al. (2014). (D and E) Comparison of chromatin accessibility changes in MPP3 cells (D) and MPP4 (E) cells to gene expression changes in GMP versus CLP cells. The y axis represents shrunken log2 fold changes of gene expression in GMP versus CLP cells. The x axis represents log2 fold changes of peak read counts in Ebf1KO versus Ebf1WT MPP3 (D) and Ebf1KO versus Ebf1WT MPP4 (E) cells. Highlighted box shows positive correlation of KO Reduced MPP3 peaks and CLP gene expression. (D and E) Left: Dots represent DA peaks in MPP3 and MPP4 cells. Right: Dots represent DE genes and ATAC peaks that overlap their TSS (TSS peaks). (F and G) GSEA results of all MPP3 (F) and MPP4 (G) ATAC peaks predicted to be enhancers (CRUP probability >0.8), ranked by log2 fold change, against a CLP-specific gene set (left) and a GMP-specific gene set (right). Positive enrichment scores reflect enrichment of the gene set in Ebf1KO cells, negative enrichment scores reflect enrichment of the gene set in Ebf1WT cells. CLP and GMP RNA-seq data analyzed in D–G was retrieved from GEO dataset GSE162662.