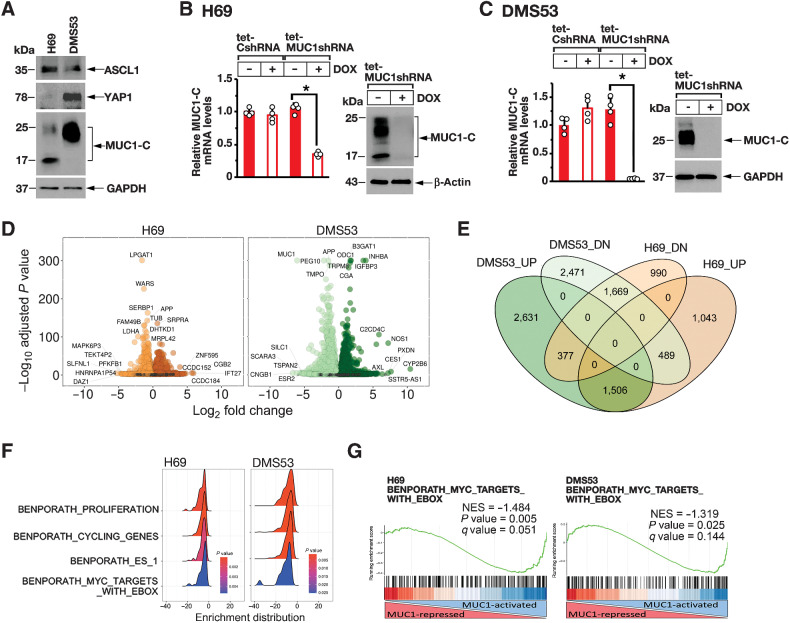
Figure 1.
MUC1-C regulates common transcriptomes in SCLC-A cells. A, Lysates from H69 and DMS53 cells were immunoblotted with antibodies against the indicated proteins. B and C. H69 (B) and DMS53 (C) cells expressing a tet-CshRNA or a tet-MUC1shRNA were treated with vehicle or 500 ng/mL DOX for 7 days. The cells were analyzed for MUC1-C mRNA levels by qRT-PCR using primers listed in Supplementary Table S1. The results (mean ± SD of four determinations) are expressed as relative mRNA levels compared with that obtained for vehicle-treated cells (assigned a value of 1; left). Lysates were immunoblotted with antibodies against the indicated proteins (right). D, RNA-seq was performed in triplicate on H69/tet-MUC1shRNA and DMS53/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days. The datasets were analyzed for effects of MUC1-C silencing on repressed and activated genes as depicted by the Volcano plots. E, Overlap of down- and upregulated genes in H69 and DMS53 cells with MUC1-C silencing. F, RNA-seq datasets from H69 (left) and DMS53 (right) cells were analyzed with GSEA for enrichment distribution using the indicated BENPORATH collection gene signatures. G, RNA-seq datasets from H69 (left) and DMS53 (right) cells were analyzed with GSEA using the BENPORATH MYC TARGETS WITH EBOX gene signatures.