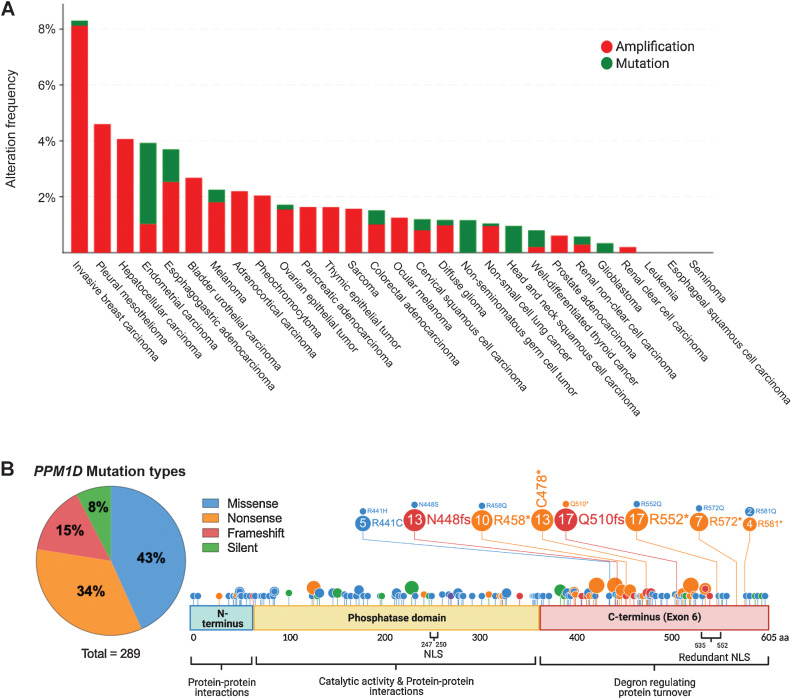
Figure 1.
PPM1D truncating mutations in solid cancers. A, Histogram showing the prevalence of PPM1D amplifications and C-terminal truncating mutations by cancer type, as indicated on the x-axis. The percentage of cases with PPM1D genomic alterations is indicated on the y-axis. The data were obtained from Pan-Cancer studies available in the cBioPortal database (10,967 total samples), which was then filtered to show only cancer types with more than 50 cases. B, Lollipop plot showing the location of the truncating mutations in the context of the domains of the PPM1D gene. A total of 289 PPM1D mutations were identified across 43 histology types from the COSMIC database. A pie chart of the mutation types is included with missense and nonsense mutations being the most common. The phosphatase domain and exon 6 of PPM1D are shown. Several mutation hotspots are noted.