Abstract
RNA interference (RNAi) is an intrinsic antiviral immune mechanism conserved in diverse eukaryotic organisms. However, the mechanism by which antiviral RNAi in mammals is regulated is poorly understood. In this study, we uncovered that the E3 ubiquitin ligase STIP1 homology and U-box-containing protein 1 (STUB1) was a new regulator of the RNAi machinery in mammals. We found that STUB1 interacted with and ubiquitinated AGO2, and targeted it for degradation in a chaperon-dependent manner. STUB1 promoted the formation of Lys48 (K48)-linked polyubiquitin chains on AGO2, and facilitated AGO2 degradation through ubiquitin-proteasome system. In addition to AGO2, STUB1 also induced the protein degradation of AGO1, AGO3 and AGO4. Further investigation revealed that STUB1 also regulated Dicer's ubiquitination via K48-linked polyubiquitin and induced the degradation of Dicer as well as its specialized form, termed antiviral Dicer (aviDicer) that expresses in mammalian stem cells. Moreover, we found that STUB1 deficiency up-regulated Dicer and AGO2, thereby enhancing the RNAi response and efficiently inhibiting viral replication in mammalian cells. Using the newborn mouse model of Enterovirus A71 (EV-A71), we confirmed that STUB1 deficiency enhanced the virus-derived siRNAs production and antiviral RNAi, which elicited a potent antiviral effect against EV-A71 infection in vivo. In summary, our findings uncovered that the E3 ubiquitin ligase STUB1 was a general regulator of the RNAi machinery by targeting Dicer, aviDicer and AGO1–4. Moreover, STUB1 regulated the RNAi response through mediating the abundance of Dicer and AGO2 during viral infection, thereby providing novel insights into the regulation of antiviral RNAi in mammals.
Keywords: Antiviral RNAi, STIP1 homology and U-box-containing protein 1 (STUB1), Argonaute 2 (AGO2), Dicer
Highlights
-
•
The E3 ubiquitin ligase, STUB1, is a novel regulator of the RNAi machinery in mammals.
-
•
STUB1 induces ubiquitination and degradation of Dicer and AGO proteins in mammals.
-
•
STUB1 deficiency enhances a potent antiviral effect against EV-A71 infection in vivo.
1. Introduction
RNAi interference (RNAi) is a conserved eukaryotic post-transcriptional gene silencing mechanism, in which small non-coding RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), mediate the gene expression. RNAi functions as an intrinsic anti-viral immunity in diverse eukaryotes. During viral infection, Dicer cleaves viral replicative intermediate double-stranded RNAs (vRI-dsRNAs) into virus-derived siRNAs (vsiRNAs) that are loaded into the RNA-induced silencing complexes (RISC) within Argonaute (AGO) protein and directs the degradation of cognate viral RNAs (Guo et al., 2019). Thus far, RNAi has been widely recognized as the major antiviral defense mechanism in fungi, plants and invertebrates, while many viruses encode viral suppressors of RNAi (VSRs) to counteract antiviral RNAi via different mechanisms.
Accumulating evidences have demonstrated that RNAi plays important antiviral roles in mammals. RNAi has been found to play efficient antiviral roles in diverse undifferentiated mammalian cells, including murine embryonic stem cells, human neural progenitor cells and neural stem cells (Maillard et al., 2013; Xu et al., 2019; Zeng et al., 2020). A very recent study has demonstrated that mouse and human stem cells encode a specialized isoform of Dicer (antiviral Dicer, aviDicer) with potent dicing activity to elicit effective antiviral RNAi response (Poirier et al., 2021; Shahrudin and Ding, 2021). Moreover, RNAi also exerts antiviral effects in differentiated somatic cells or mice against a wide range of RNA viruses, while these viruses encode VSRs that are required for replication and act to inhibit the vRI-dsRNAs processing by Dicer (Han et al., 2020b; Li et al., 2013, 2016; Qian et al., 2020; Qiu et al., 2017, 2020; Zhang et al., 2020b). Interestingly, we have demonstrated that the potency of antiviral RNAi can be successfully unlocked by abrogating VSRs via rationally designed peptides, which highlight the physiological and functional importance of antiviral RNAi in mammals (Fang et al., 2021; Sanchez-David and Maillard, 2021).
Dicer and AGO family proteins are the critical components of the RNAi machinery. Mammals encode a single Dicer that processes both pre-miRNAs and long dsRNAs, including vRI-dsRNAs. On the other hand, mammals possess four AGO proteins, AGO1–AGO4. And AGO2 is the only catalytically active AGO that is required for both the miRNA-mediated gene silencing and the vsiRNA-mediated antiviral RNAi. Interestingly, a recent study has shown that mammalian AGO4 also possesses antiviral activity both dependently and independently of type I interferon (IFN-I) (Adiliaghdam et al., 2020).
Homeostatic mechanisms orchestrate the abundance of the components in the RNAi machinery and their functions in mammals. Multiple lines of evidence have shown that the ubiquitin-proteasome system (UPS) controls the stability and turnover of Dicer and AGO2, and this regulation plays important role in modulating miRNA expression profiles and RISC-mediated gene silencing in a temporal or cell state-dependent manner (Bronevetsky et al., 2013; Smibert et al., 2013). To date, only a handful of E3 ubiquitin ligases targeting Dicer and/or AGO2 for proteasomal degradation have been identified. The ubiquitin ligase Trim71 (or mLin41), a known stem cell factor, has been found to ubiquitinate AGO2 and Dicer, and regulate AGO2 protein level in mouse embryonic stem cells (Rybak et al., 2009). Recent studies have demonstrated that the E3 ubiquitin ligase ZSWIM8 elicits AGO2 proteasomal degradation when its miRNA guide binds a highly complementary target RNA, a process called target-directed miRNA degradation (TDMD) (Han et al., 2020a; Shi et al., 2020). Moreover, the E3 ubiquitin ligase STIP1 homology and U-box-containing protein 1 (STUB1), also known as the C-terminus of HSC70-interacting protein (CHIP), has recently been identified to participate in the process of Dicer degradation induced by oxaliplatin, a chemotherapeutic agent, in cancer cells (Lai et al., 2018), but whether STUB1 directly regulates Dicer degradation remain unclear. Therefore, although certain ubiquitin ligases have been implicated in the turnover of Dicer and AGO2, the molecular mechanism leading to their down-regulation is still less understood.
Moreover, previous studies on the proteasomal degradation of Dicer and/or AGO2 have focused mostly on the controls of miRNA biogenesis and miRNA-directed gene silencing. For example, Trim71 facilitates the degradation of AGO2 and interferes with let-7- and miR-124-mediated gene silencing in mouse stem cells, which plays important role in regulating stem cell development (Liu et al., 2021; Rybak et al., 2009). Besides, AGO2 has been found to be inducibly ubiquitinated in activated T cells and the reduced AGO2, as well as miRNA expressions in naïve T cells differentiate more readily into cytokine-producing helper T cells (Bronevetsky et al., 2013). However, whether and how regulating the abundance of Dicer and AGO2 affects the RNAi-directed antiviral activity in mammals remains elusive.
In this study, we identified that STUB1 was a new regulator for the RNAi machinery in mammalian cells. STUB1 interacted with AGO2, promoted the formation of K48-linked polyubiquitin chains on AGO2 and induced proteasomal degradation. Further investigation uncovered that STUB1 also mediated the protein expression levels of the other three AGO. Moreover, STUB1 directly interacted with Dicer and targeted it for K48-linked polyubiquitin formation and degradation. Interestingly, the Dicer's specialized form, aviDicer, could also be regulated by STUB1. Moreover, we provided evidence that STUB1 mediated antiviral RNAi by regulating the abundance of Dicer and AGO2 during viral infection in cells and in vivo.
2. Materials and methods
2.1. Cell culture and reagents
HEK293T (ATCC, CRL-11268), RD (CCL-136), A549 (CCL-185) and Vero (CCL-81) cells were cultured with Dulbecco's modified Eagle's medium (DMEM, Gibco, Thermo-Fisher, Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco, Thermo-Fisher, Waltham, MA, USA), 100 U/mL penicillin/streptomycin at 37 °C in an incubator with 5% CO2. Cells grown to approximately 80% confluence in the Petri dish with a glass bottom (NEST, Wuxi, China) were transfected with plasmid by using Lipofectamine 2000 Reagent (Life Technologies, Thermo-Fisher, Waltham, MA, USA). DMSO, 3-methyladenine (3-MA), Bafilomycin A1 (Baf-A1), chloroquine, MG132 was from Sigma (Sigma-Aldrich, Milwaukee, USA) and Ruxolitinib was from Selleck (Houston, TX, USA).
2.2. Plasmids, siRNA and virus
FLAG-tagged human AGO2, FLAG-tagged human Dicer, His-tagged STUB1 and their truncations were cloned into pRK and verified by Sanger sequencing (Tsingke, China). EGFP and shEGFP cloned into pUC57 were prepared in our laboratory (Qiu et al., 2017). The siRNAs targeting EGFP were synthesized by RiboBio (Guangzhou, China). EV-A71 strain H (VR-1432) and EV-A71D23A were prepared as previously described (Qiu et al., 2017).
2.3. Co-immunoprecipitation, western blotting and antibodies
Co-immunoprecipitation and Western blotting were conducted as previously described with minor modifications (Xu et al., 2021). In brief, cell monolayers were washed with PBS and incubated on ice with lysis buffer containing 20 mmol/L Tris-HCl, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP-40 and 1% protease and phosphatase inhibitor cocktail (Roche, Shanghai, China). For each sample, 600 μL protein lysate was incubated with 1 μg antibody and 30 μL protein A/G magnetic beads (MCE, Shanghai, China) overnight at 4 °C. The magnetic beads were washed three times with 1 mL of lysis buffer, and then the precipitates were detected by Western blotting.
For Western blotting, cells were harvested and lysed as described above. Equal amounts of protein samples were denatured for 15 min in 5 × SDS-PAGE loading buffer (Beyotime, Shanghai, China). Proteins were separated on SDS-PAGE gels and then electro-transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA), which were then blocked for 1 h at 25 °C in Tris-buffered saline (SigmaAldrich) containing 5% nonfat milk powder and 0.1% Tween 20 (Sigma-Aldrich). Next, the membranes were incubated with primary antibodies at 4 °C overnight and then with the corresponding secondary antibodies conjugated to horseradish peroxidase at room temperature for 1 h. The protein bands were detected using the ECL kit.
The antibodies used are as follows: anti-FLAG (Sigma-Aldrich), anti-HA (Abclone, Wuhan, China), anti-pan-AGO (Millipore), anti-His and anti-STUB1 were from Proteintech (Wuhan, China), anti-Dicer were from Abcam (Cambridge, UK).
2.4. Quantitative real-time PCR
Total RNAs were extracted using Trizol reagent (TAKARA, Dalian, China) according to the manufacturer's instructions. cDNA was acquired by using SuperScript II Reverse Transcriptase (TAKARA). cDNA was diluted in nuclease-free water, and gene expression was analyzed by qPCR SYBR Green Master Mix (Yeasen, Shanghai, China) with ABI 7500 FAST machine (Applied Biosystems) as described previously (Fang et al., 2021; Qiu et al., 2017, 2020). Differences between experimental and control groups were tested using Student's t-test. P values less than 0.05 were considered statistically significant.
2.5. Lentiviral shRNA packaging, infection and selection
For packaging lentiviruses, 1 μg pLKO.1 shRNA plasmid (Addgene), 750 ng psPA × 2 packaging plasmid (Addgene), and 250 ng pMD2.G envelope plasmid (Addgene) were co-transfected into 7 × 104 HEK293T cells with 6 μL Lipofectamine 2000 Reagent (Life Technologies). The supernatants containing lentiviruses were collected, filtered and stored at −80 °C. For infection, HEK293T, A549, RD and Vero cells were incubated with viral stocks supplemented with 8 mg/mL polybrene (Solarbio, Beijing, China) for 10 h, and then supplied with fresh medium. Cells were selected with puromycin (Invitrogen) at 24 h post-infection (h.p.i.). The following primers were used: human STUB1 shRNA: GAAGCGCTGGAACAGCATTGA; mouse STUB1 shRNA: GGAGATGGAGAGTTATGATGA. The human STUB1 shRNA was also used to knock down STUB1 in Vero cells.
2.6. Mouse experiments
Mice were specific-pathogen free and kept under a 12 h light/dark cycle with a standard chow diet. All animal experiments were performed in strict accordance with the guidelines of the Chinese Regulations of Laboratory Animals (Ministry of Science and Technology of China) and approved by the Institutional Animal Care and Use Committee at Wuhan Institute of Virology, Chinese Academy of Sciences. EV-A71-infected newborn mice were established as previously described (Fang et al., 2021). Briefly, we injected lentivirus expressing shSTUB1 or shScramble into the hind leg muscles of newborn specific-pathogen-free (SPF) ICR mice (Experimental Animal Center, Wuhan Institute of Virology). Two days later, mice were i.p. infected with EV-A71 (106 PFU/per mouse). At 4 days post-infection (d.p.i.), mice were euthanized and the hind leg muscles were harvested for subsequent analysis.
2.7. Deep small RNA sequencing
The deep sequencing and data analysis have been described previously (Fang et al., 2021). Briefly, libraries of small RNAs were constructed using TruSeq Small RNA Library Preparation Kits (Illumina) according to manufacturer's protocols, and then sequenced by Illumina HiSeq (2000) at Bioacme (Wuhan, China). The sequenced reads were mapped to the mouse genome (release GRCm38) by using Bowite 2.2.5. The vsRNA reads were mapped to EV-A71 genome with 100% identity. The distribution and length of vsRNA reads were conducted by using in-house Perl scripts and Excel.
3. Results
3.1. STUB1 interacts with AGO2 in human cells
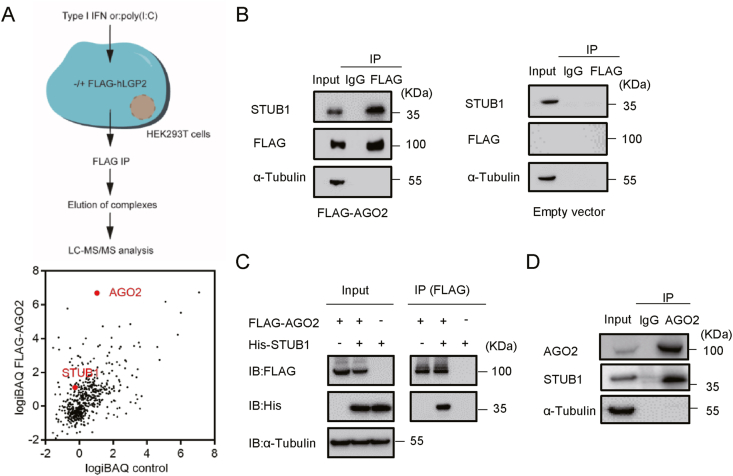
To determine the potential host factors regulating AGO2, we ectopically expressed FLAG-tagged human AGO2 in cultured HEK293T cells, and performed co-immunoprecipitation assay using anti-FLAG antibody. The immunoprecipitates were then analyzed via LC-MS/MS (Fig. 1A). Among the interactors of AGO2, we identified STUB1, a chaperone-associated E3 ubiquitin ligase (Fig. 1B). To verify the AGO2-STUB1 interaction, co-immunoprecipitation assays were conducted by using HEK293T cells expressing FLAG-AGO2 together with His-tagged human STUB1. We found that STUB1, but not the control vector, was associated with AGO2 in cells (Fig. 1C). Moreover, the endogenous AGO-STUB1 interaction was further confirmed via co-immunoprecipitation in HEK293T cells with anti-pan AGO antibody (Fig. 1D). Together, our results uncovered that STUB1 was a binding partner of AGO2 in human cells.
Fig. 1.
STUB1 interacts with AGO2 in mammalian cells. A Schematic illustration of the experimental approach used to identify the interactors of AGO2. HEK293T cells were transfected with the FLAG-tagged human AGO2 plasmid or empty vector (control) for 24 h, and then lysed and subjected to co-immunoprecipitation by using anti-FLAG antibody. The AGO2-bound complexes were excised and analyzed by LC-MS/MS. Graphs display the intensity-based absolute quantification (iBAQ) score of proteins identified in the FLAG-AGO2-immunoprecipitated complex versus the control. AGO2 and STUB1 were highlighted. B Western blotting analysis of the immunoprecipitated complexes as described in A. C HEK293T cells were co-transfected with FLAG-tagged human AGO2 plasmids and His-tagged human STUB1 plasmids or empty plasmid for 24 h, and subjected to co-immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-FLAG, anti-His and anti-α-Tubulin antibodies, respectively. D HEK293T cells were subjected to endogenous co-immunoprecipitation with anti-pan AGO antibody, followed by immunoblotting with anti-AGO2, anti-STUB1 and anti-α-Tubulin antibodies, respectively.
3.2. The STUB1 TPR domain and AGO2 PIWI domain are important for STUB1-AGO2 interaction
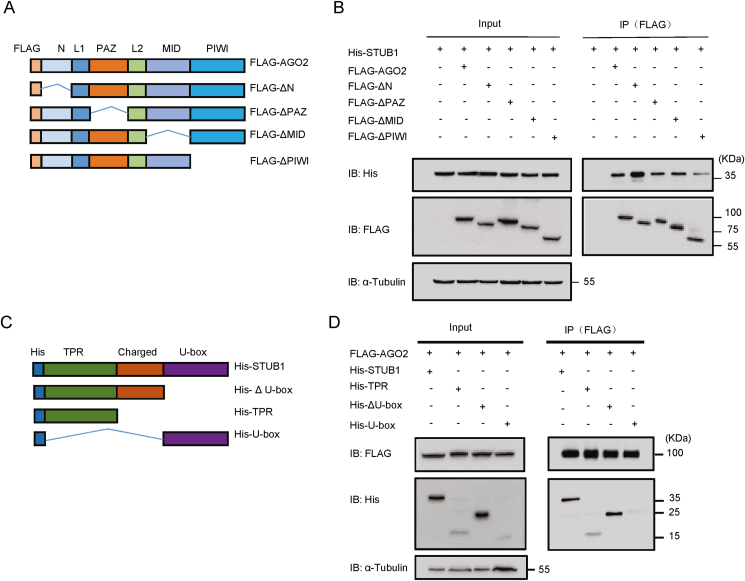
AGO2 consists of N-terminal (N), PAZ (PIWI/AGO/Zwille), MID (middle), and PIWI (P-element-induced whimpy testes) domains. To further characterize the interaction between AGO2 and STUB1, we constructed a series of AGO2 truncations (Fig. 2A) and expressed them together with His-STUB1 in HEK293T cells. Domain mapping analysis showed that the interaction between STUB1 and AGO2 was substantially reduced when the PIWI domain was deleted from AGO2, suggesting that the PIWI domain was critical for AGO2 interacting with STUB1 (Fig. 2B).
Fig. 2.
The STUB1 TPR domain and AGO2 PIWI domain are indispensable for STUB1-AGO2 interaction. A Schematic illustration of domains for the FLAG-tagged full-length AGO2 and its truncations. B HEK293T cells were co-transfected with His-STUB1 together with FLAG-AGO2 or its truncations for 24 h, and then subjected to co-immunoprecipitation with anti-FLAG antibody. C Schematic illustration of domains for the His-tagged full-length STUB1 and its truncations. D HEK293T cells were co-transfected with FLAG-AGO2 together with His-STUB1 or its truncations for 24 h, and then subjected to co-immunoprecipitation with anti-FLAG antibody.
We sought to identify the region of STUB1 responsible for its interaction with AGO2. STUB1 consists of three tetratricopeptide repeats (TPR) domain at its N-terminal, a charged domain in the middle and a U-box domain at its C-terminal. We then constructed a series of STUB1 truncations (Fig. 2C) and expressed them with FLAG-AGO2 in HEK293T cells. As shown in Fig. 2D, His-U-box (deleting TPR domain) failed to interact with AGO2, while His-ΔU-box or His-TPR (both of them containing TPR domain) still interacted with AGO2, suggesting that the TPR domain was important for STUB1 to interact with AGO2. Together, these data suggested that STUB1 TPR domain and AGO2 PIWI domain were important for STUB1-AGO2 interaction.
3.3. STUB1 promotes the ubiquitination and degradation of AGO2 dependently on its E3 ligase and chaperon-binding activities
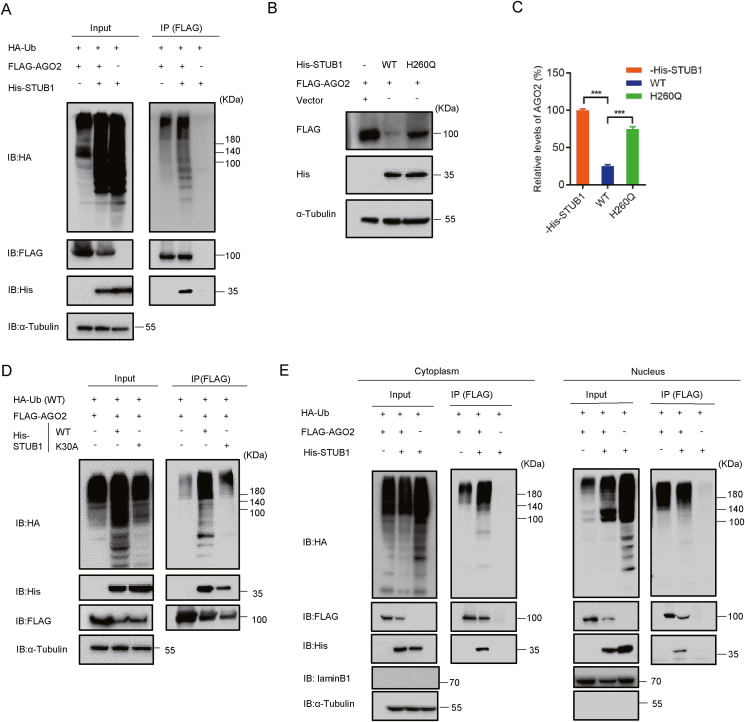
STUB1 is an E3 ligase that can mediate ubiquitination and degrade different substrates through the proteasomal pathway (Zhang et al., 2020a). Therefore, we examined whether AGO2 is also the target for STUB1-mediated ubiquitination and degradation. To this end, HEK293T cells were transfected with plasmids for FLAG-AGO2, His-STUB1 and HA-ubiquitination (HA-Ub), and then subjected to co-immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA antibody to detect ubiquitinated-AGO2. Our data showed that overexpression of STUB1 dramatically enhanced AGO2 ubiquitination and reduced the protein level of AGO2 in cells (Fig. 3A).
Fig. 3.
STUB1 promotes the ubiquitination and degradation of AGO2 dependently on its E3 ligase and chaperon-binding activities. A HEK293T cells were co-transfected with HA-Ub together with FLAG-AGO2 and/or His-STUB1 for 24 h, respectively, and subjected to co-immunoprecipitation with anti-FLAG antibody. The ubiquitinated AGO2 was detected with anti-HA antibody, and AGO2-bound STUB1 was detected with anti-His antibody. B–C HEK293T cells were co-transfected with FLAG-AGO2 together with wild-type (WT) STUB1, STUB1H260Q mutant or empty plasmid, respectively, followed by immunoblotting with anti-FLAG antibody to detect AGO2. The experiments were repeated three times, and means ± S.D. were plotted. Student's t-test was used. ∗∗∗, P < 0.001. D HEK293T cells were co-transfected with HA-Ub together with FLAG-AGO2 and His-STUB1 or His-STUB1K30A, respectively, followed by immunoblotting with anti-HA antibody to detect ubiquitinated AGO2. E HEK293T cells were co-transfected with HA-Ub together with FLAG-AGO2 and/or His-STUB1 for 24 h, respectively. The Cellular nuclear and cytoplasmic fractions were separated and were immunoprecipitated with anti-FLAG antibody, followed by immunoblotting with anti-HA antibody to detect ubiquitinated AGO2. LaminB1 and tubulin were used as the nuclear and cytoplasmic makers, respectively.
We then investigated whether the E3 ligase activity of STUB1 was required for the ubiquitination of AGO2. Previous study reported that the residue H260 within the Ubox domain was required for the E3 ubiquitin ligase activity of STUB1, as a single-point of this residue (i.e. H260Q) made STUB1 incapable to ubiquitinate substrates and induce Ub-dependent degradation (Ullah et al., 2020). Our results showed that unlike wild-type (WT) STUB1, the catalytically inactive STUB1H260Q failed to affect the protein level of AGO2 (Fig. 3B and C).
STUB1 is a chaperone-associated E3 ubiquitin ligase. Previous study has shown that single-point K30A mutation in the TPR domain abolishes the binding of STUB1 to chaperones (Hsc70 or Hsp90), which results in the attenuated STUB1-substrate interaction as well as the reduced ubiquitination of substrates (Yonezawa et al., 2017). Our co-immunoprecipitation assays uncovered that the ubiquitination levels of AGO2 were substantially reduced in the presence of STUB1K30A when being compared with that of wild-type STUB1 (Fig. 3D), suggesting that the STUB1-chaperone interaction was required for STUB1 mediating the ubiquitination of AGO2.
AGO2 has been identified to be localized in both the cytoplasm and nucleus (Gebert and MacRae, 2019). Moreover, a previous study showed that STUB1 can also translocate from cytoplasm to nuclear (Yonezawa et al., 2017). Thus, to examine whether STUB1 could regulate the ubiquitination and degradation of AGO2 in the nucleus, we isolated the nuclear and cytoplasmic fractions of HEK293T cells expressing HA-Ub together with FLAG-AGO2 and/or His-STUB1. The isolated fractions were then subjected to co-immunoprecipitation assays with anti-FLAG antibody. Our results showed that STUB1 interacted with AGO2 and induced AGO2 ubiquitination/degradation in both cytoplasm and nucleus (Fig. 3E).
Together, our findings indicated that STUB1 ubiquitinated AGO2 and targeted it for degradation, and the E3 ligase and chaperone-binding activities were required for STUB1 to regulate AGO2 ubiquitination.
3.4. STUB1 down-regulates the protein level of AGO2 through the ubiquitin-proteasome system
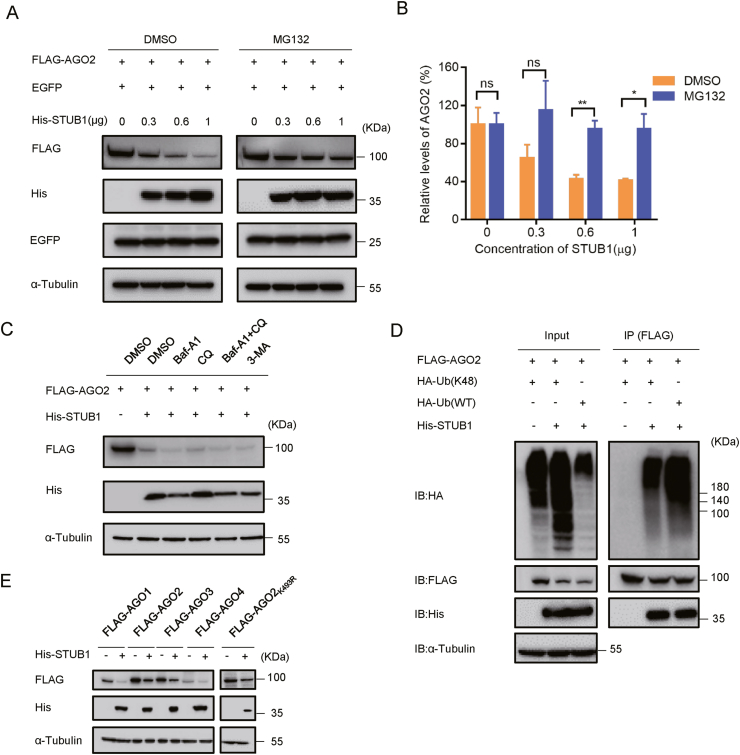
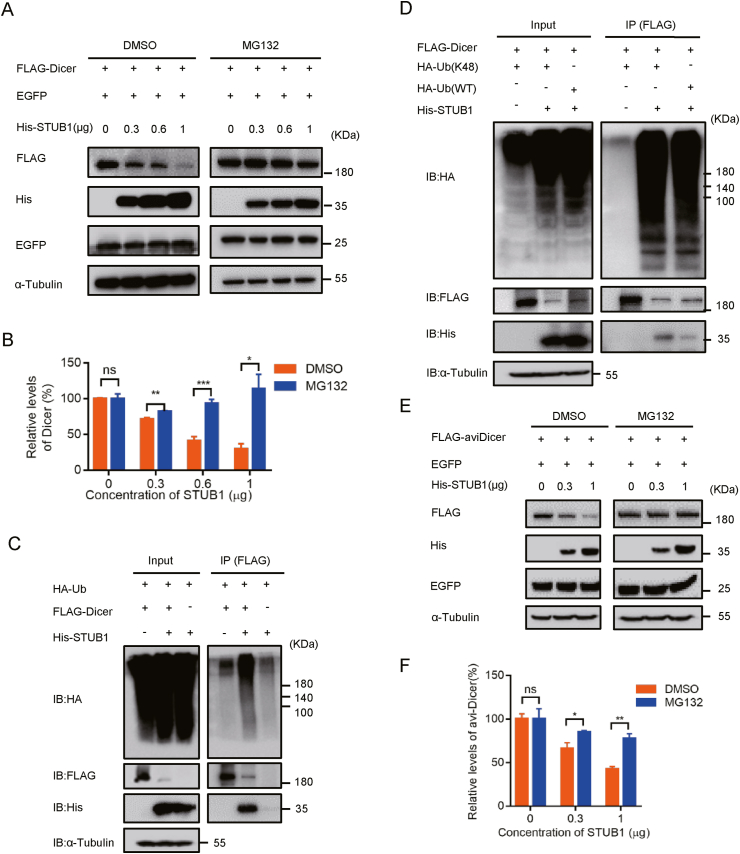
After finding that STUB1 regulated AGO2 ubiquitination and degradation, we sought to examine whether the ubiquitin-proteasome system (UPS) was essential for the degradation of AGO2. To this end, we examined the AGO2 protein level in cells transfected with FLAG-AGO2 and His-STUB1 in the presence of MG132, a proteasome inhibitor. Our results showed that STUB1 substantially reduced the protein level of AGO2 in a dose-dependent manner in DMSO-treated cells, while the AGO2 protein level was stable in cells treated with MG132 (Fig. 4A and B). EGFP was used as the reference in this experiment, and its protein level was nearly not affected in the presence of either STUB1 or MG132, showing that STUB1 specifically targeted AGO2. These results indicated that STUB1 regulated AGO2 degradation through UPS.
Fig. 4.
STUB1 down-regulates the AGO2 protein level through the ubiquitin-proteasome system. A–B HEK293T cells were co-transfected FLAG-AGO2 together with EGFP and different concentrations of His-STUB1, respectively. After 24 h transfection, cells were treated with DMSO or MG132 (10 μmol/L) for 9 h. The resulting cell lysates were immunoblotted with the indicated antibodies. The experiments were repeated three times, and means ± S.D. were plotted. Student's t-test was used. ∗, P < 0.05; ∗∗, P < 0.01; ns, no significant. C HEK293T cells were co-transfected with FLAG-AGO2 together with His-STUB1 or empty vector for 24 h, and then treated with DMSO, 20 μmol/L chloroquine (CQ), 100 nmol/L Bafilomycin A1 (Baf-A1), 10 μmol/L 3-methyladenine (3-MA) or 100 nmol/L Baf-A1 together with 10 μmol/L 3-MA. After 24 h treatment, the resulting cell lysates were immunoblotted with the indicated antibodies. D HEK293T cells were co-transfected with FLAG-AGO2 together with His-STUB1 and WT Ub or K48 Ub for 24 h, respectively, and subjected to co-immunoprecipitation with anti-FLAG antibody, and followed by immunoblotting with anti-HA antibody to detect ubiquitinated AGO2. E HEK293T cells were co-transfected with FLAG-AGO1, FLAG-AGO2, FLAG-AGO3, FLAG-AGO4 or FLAG-AGO2K493R together with His-STUB1 or empty plasmid for 24 h, respectively, and subjected to western blotting with anti-FLAG antibody to detect AGO proteins.
Moreover, we examined whether the AGO2 degradation by STUB1 can be mediated through autophagy by using autophagosome-lysosome inhibitors, including 3-methyladenine (3-MA), chloroquine (CQ), and bafilomycin A1 (Baf A1) (Sun et al., 2018). As shown in Fig. 4C, none of these inhibitors could restore the AGO2 protein level in STUB1-expressing cells, showing that the AGO2 degradation mediated by STUB1 was irrespective of autophagy. Moreover, it is well-known that Lys48 (K48)-linked Ub mediates the proteasomal degradation of ubiquitinated substrates. We found that overexpression of K48-linked Ub induced AGO2 ubiquitination as efficiently as wild-type Ub did (Fig. 4D), indicating that STUB1 promotes the formation of K48–linked polyubiquitin chains on AGO2.
After finding that STUB1 regulates the proteasomal degradation of AGO2, it would be intriguing to examine whether STUB1 would also target the other three AGO proteins, as AGO1–4 share a conserved structural architecture. Thus, HEK293T cells were co-transfected with FLAG-tagged human AGO1, AGO2, AGO3 or AGO4 together with His-STUB1 or empty vector, respectively. Our data showed that the protein levels of all the four AGO proteins were remarkably reduced in cells expressing His-STUB1 (Fig. 4E). These results indicate that STUB1 can target AGO1–4 for degradation. Moreover, previous studies have demonstrated that ZSWIM8, an ubiquitin E3 ligase, promotes the TDMD-induced proteasomal degradation of AGO2 in mammals (Han et al., 2020a; Shi et al., 2020). To exclude the possibility that STUB1-mediated AGO2 degradation is relevant to ZSWIM8, we constructed an AGO2K493R mutant, which has been shown to be irresponsive to ZSWIM8 (Han et al., 2020a). Our results showed that overexpression of STUB1 reduced the protein level of AGO2K493R (Fig. 4E), confirming that STUB1-mediated AGO2 degradation is irrespective of ZSWIM8 and TDMD.
Together, our findings indicated that STUB1 mediated AGO2 ubiquitination for proteasomal degradation. Moreover, STUB1 can also induce the degradation of the other three AGO proteins. Because AGO2 is the main effector of the RNAi pathway, we focused on the STUB1-mediated AGO2 degradation in the subsequent experiments.
3.5. STUB1 regulates Dicer ubiquitination for proteasomal degradation
After establishing the proteasomal degradation of AGO2 by STUB1, we examined whether STUB1 would affect the abundance of Dicer. The protein level of Dicer was substantially down-regulated in STUB1-expressing cells treated with DMSO, while MG132 treatment restored the reduced protein level of Dicer (Fig. 5A and B). Moreover, co-immunoprecipitation assay by using cells expressing HA-Ub together with FLAG-Dicer and His-STUB1 confirmed that STUB1 interacted with Dicer, and targeted it for ubiquitination and degradation (Fig. 5C). Besides, we also showed that STUB1 induced the K48–linked polyubiquitin on Dicer (Fig. 5D).
Fig. 5.
STUB1 regulates Dicer ubiquitination for proteasomal degradation. A-B HEK293T cells were co-transfected FLAG-Dicer together with EGFP and different concentrations of His-STUB1, respectively. After 24 h transfection, cells were treated with DMSO or MG132 (10 μmol/L) for 9 h. The resulting cell lysates were immunoblotted with the indicated antibodies. The experiments were repeated three times, and means ± S.D. were plotted. Student's t-test was used. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ns, no significant. C HEK293T cells were co-transfected with HA-Ub together with FLAG-Dicer and/or His-STUB1 for 24 h, respectively, and subjected to co-immunoprecipitation with anti-FLAG antibody. The ubiquitinated AGO2 was detected with anti-HA antibody, and Dicer-bound STUB1 was detected with anti-His antibody. D HEK293T cells were co-transfected with FLAG-Dicer together with His-STUB1 and WT Ub or K48-Ub for 24 h, respectively, and subjected to co-immunoprecipitation with anti-FLAG antibody, and followed by immunoblotting with anti-HA antibody to detect ubiquitinated Dicer. E–F HEK293T cells were co-transfected FLAG-aviDicer together with EGFP and different concentrations of His-STUB1, respectively. After 24 h transfection, cells were treated with DMSO or MG132 (10 μmol/L) for 9 h. The resulting cell lysates were immunoblotted with the indicated antibodies. Student's t-test was used. ∗, P < 0.05; ∗∗, P < 0.01; ns, no significant.
Moreover, a recent study has shown that human and mouse stem cells transcribe an alternatively spliced Dicer mRNA isoform that encodes a specialized aviDicer protein absent of N-terminal Hel2i domain and elicits a potent antiviral RNAi response (Poirier et al., 2021). It is intriguing to examine whether STUB1 would also mediate the degradation of aviDicer. Thus, we constructed the aviDicer mutant and examined its protein level in the presence or absence of STUB1 in HEK293T cells with or without MG132 treatment. Our results showed that STUB1 substantially reduced aviDicer protein level in a dose-dependent manner in DMSO-treated but not in MG132-treated cells (Fig. 5E and F). These results indicate that STUB1 mediates the degradation of both Dicer and aviDicer via UPS in mammalian cells.
3.6. STUB1 regulates the RNAi response through mediating the abundance of Dicer and AGO2 in mammalian cells
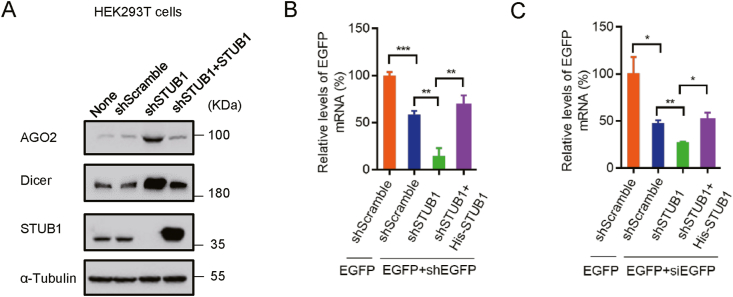
We sought to investigate whether STUB1 could mediate the activity of the RNAi machinery by regulating AGO2 and Dicer. To this end, we constructed a HEK293T cell line with stable STUB1 knockdown via lentiviral shRNA targeting human STUB1 (shSTUB1). A stable cell line expressing scramble shRNA (shScramble) was established and used as the negative control. The protein levels of AGO2 and Dicer were remarkably increased in shSTUB1 cells when being compared with that in shScramble or mock cells, while overexpressing His-STUB1 counterbalanced the effect of STUB1 knockdown on the protein levels of AGO2 and Dicer (Fig. 6A). After that, shScramble or shSTUB1 cells were co-transfected with the plasmids encoding EGFP and EGFP-specific shRNA (shEGFP) or EGFP-specific siRNA (siRNA), respectively. As expected, the mRNA level of EGFP was significantly reduced in shScramble cells (Fig. 6B and C), in which RNAi was induced by shEGFP or siEGFP to destruct EGFP mRNA. Moreover, the EGFP mRNA levels were further decreased in shSTUB1 cells (Fig. 6B and C), showing that STUB1 knockdown enhanced both the shRNA- and siRNA-induced RNAi. On the other hand, the levels of EGFP mRNA were significantly enhanced in shSTUB1 cells overexpressing FLAG-STUB1 compared with those in shSTUB1 cells (Fig. 6B and C), indicating that the down-regulation of Dicer and AGO2 reversed the enhanced RNAi response in shSTUB1 cells. Therefore, we concluded that STUB1 could inhibit either the shRNA- or siRNA-induced RNAi response by mediating the protein levels of Dicer and AGO2.
Fig. 6.
STUB1 regulates the RNAi response through mediating the abundance of Dicer and AGO2 in mammalian cells. A HEK293T cells were transfected with lentivirus-based shScramble or shSTUB1. The stable STUB1 knocking-down cells were examined via Western blotting with anti- AGO2, anti-Dicer and anti-STUB1 antibodies, respectively. The stable STUB1 knocking-down cells were transfected with His-STUB1 for 24 h to generate the STUB1 expression restored cells (shSTUB1+STUB1). B–C The shScramble, shSTUB1 and shSTUB1+STUB1 cells were co-transfected with EGFP together with EGFP-specific shRNA (shEGFP) or siEGFP for 24 h, and EGFP mRNA levels were examined by qRT-PCR. The experiments were repeated three times, and means ± S.D. were plotted. Student's t-test was used. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
3.7. STUB1 deficiency inhibits the replication of VSR-deficient enterovirus A71 (EV-A71D23A) through up-regulating Dicer and AGO2 in mammalian cells
It would be intriguing to examine whether STUB1, as a negative regulator of the RNAi machinery, had any functional relevance to viral infection. Previous studies by others and us have shown that the antiviral RNAi response is more evident in differentiated mammalian somatic cells when being infected with VSR-disabled mutant viruses (Han et al., 2020b; Li et al., 2013, 2016; Qian et al., 2020; Qiu et al., 2017, 2020; Zhang et al., 2020b). Thus, we used EV-A71D23A, a VSR-deficient EV-A71 that has been previously engineered by us and is sensitive to antiviral RNAi (Qiu et al., 2017), in the subsequent experiments. STUB1 deficiency in EV-A71D23A-infected cells would inhibit the replication of this mutant virus as antiviral RNAi is enhanced when the protein levels of AGO2 and Dicer are up-regulated due to the loss of STUB1.
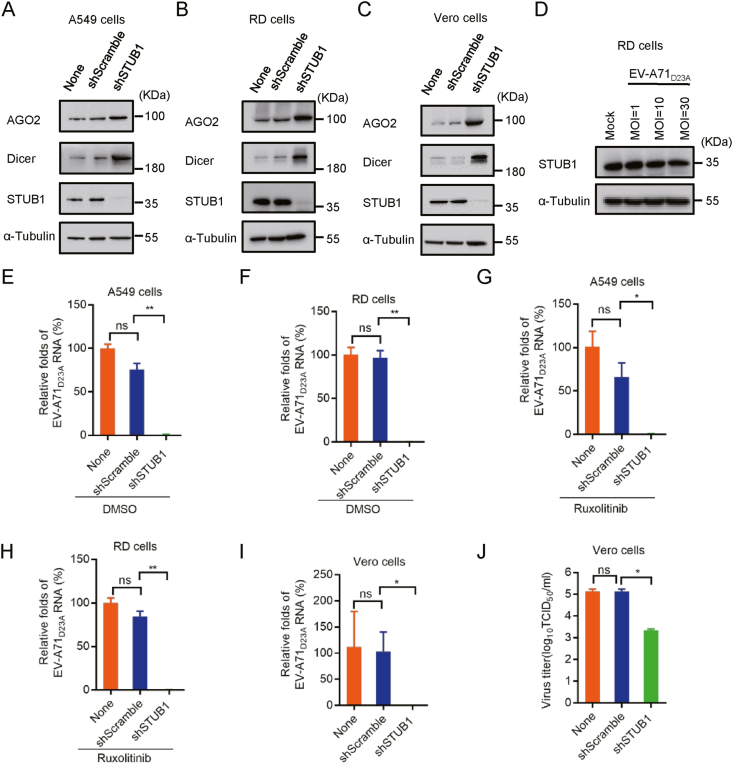
The stable STUB1 knockdown A549, RD and Vero cell lines were constructed based on lentivirus, and as expected, the deficiency of STUB1 resulted in increased protein levels of both Dicer and AGO2 in these cells (Fig. 7A–C). To exclude the potential inference of IFN-I system, we treated A549 and RD cells with or without Ruxolitinib, a JAK1 and JAK3 inhibitor to block IFN-I signaling, prior to EV-A71D23A infection at a multiplicity of infection (MOI) of 1. Besides, we showed that viral infection had no or little effect on the expression level of STUB1 at 24 h.p.i. (Fig. 7D). The viral RNA accumulation level of EV-A71D23A was remarkably reduced in STUB1-deficient cells compared to that in mock cells or shScramble cells at 24 h.p.i. (Fig. 7E and F). Moreover, the inhibitory effect of STUB1 deficiency on EV-A71D23A replication was not affected by Ruxolitinib treatment (Fig. 7G and H). The inhibitory effect of STUB1 deficiency on EV-A71D23A replication was further examined in STUB1 knocking-down Vero cells, which were defective in IFN-I incompetent system, via measuring the viral RNA accumulation and titers, respectively (Fig. 7I and J).
Fig. 7.
STUB1 deficiency inhibits the replication of VSR-deficient Enterovirus A71 (EV-A71 D23A) through up-regulating Dicer and AGO2 in mammalian cells. A-C A549, RD and Vero cells were transfected with lentivirus-based shScramble or shSTUB1, respectively. Cells without lentivirus treatment were used as controls (None). The stable STUB1 knocking-down cells were examined via Western blotting with anti- AGO2, anti-Dicer and anti-STUB1 antibodies, respectively. D The protein levels of STUB1 were examined in RD cells infected with different MOI EV-A71D23A at 24 h.p.i. E–J The indicated A549, RD and Vero cells were used to infected with EV-A71D23A at an MOI of 1 treated with DMSO or Ruxolitinib (2 μmol/L), respectively. At 24 h.p.i., the levels of EV-A71D23A genomic RNAs were detected by qRT-PCR. The viral titers in infected Vero cells were examined. TCID50, 50% tissue culture infectious doses per milliliter. The experiments were repeated three times, and means ± S.D. were plotted. Student's t-test was used. ∗, P < 0.05; ∗∗, P < 0.01.
Together, our findings showed that loss of STUB1 strengthened antiviral RNAi, thereby inhibiting the replication of VSR-deficient virus in an IFN-I irrespective manner in mammalian cells.
3.8. STUB1 deficiency enhances antiviral RNAi response in vivo
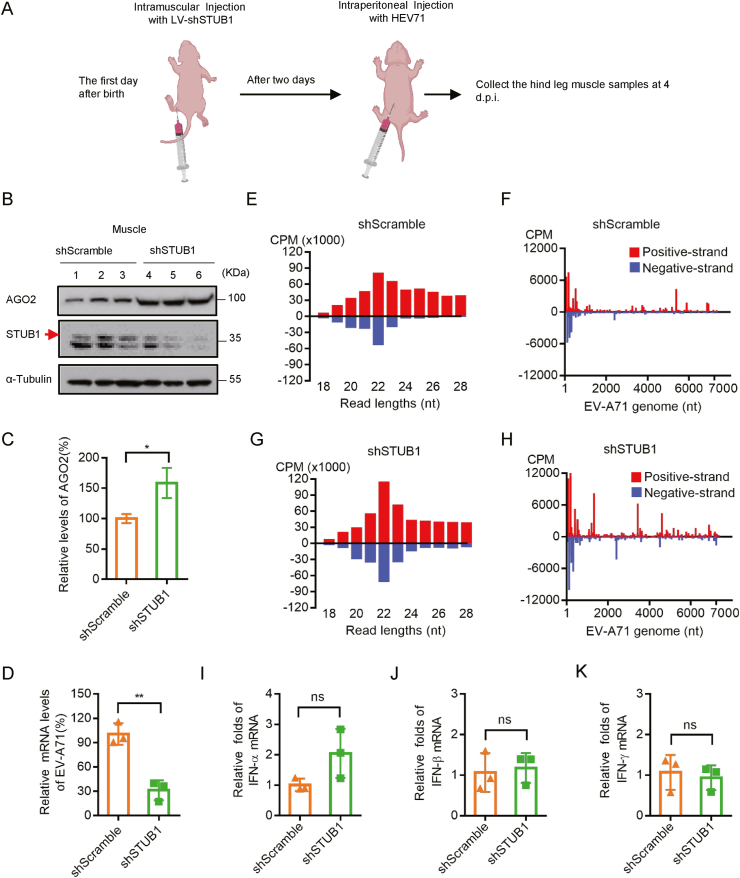
We sought to examine the physiological importance of STUB1 in mediating antiviral RNAi in vivo. Previous studies by others and us have shown that infection by Nodamura virus, EV-A71 or Zika virus can result in the production of vsiRNAs in suckling mice, indicating that newborn mouse is a good model to study antiviral RNAi (Fang et al., 2021; Han et al., 2020b; Li et al., 2013; Zhang et al., 2020b, 2021). Therefore, the roles of STUB1 in regulating the RNAi response were examined in this model. Thus, we injected lentivirus expressing murine shSTUB1 or shScramble into the hind leg muscles of ICR newborn mice to generate STUB1 knockdown or scramble mice, respectively (Fig. 8A). Western blotting analysis showed that the protein level of murine AGO2 was correspondingly upregulated in shSTUB1 mice (Fig. 8B and C), indicating the regulation of the RNAi machinery by STUB1 is conserved in mammals. Subsequently, shSTUB1 and shScramble mice were infected with 1 × 106 PFU of WT EV-A71 by intraperitoneal (i.p) injection.
Fig. 8.
STUB1 deficiency enhances antiviral RNAi response in vivo. A The lentivirus-based shScramble or mouse shSTUB1 was intramuscularly injected into the hind leg muscles of 1-day-old ICR mice (n = 3 for each group). Two days later, mice were then i.p. injected with 1 × 106 PFU EV-A71. At 4 d.p.i., mice were euthanized and the hind leg muscles were harvested for subsequent analysis. B The expression levels of mouse AGO2 and STUB1 in shScramble and shSTUB1 mice were tested by Western blotting. The resulting cell lysates were immunoblotted with the indicated antibodies. C The relative expression levels of AGO2 in shScramble and shSTUB1 mice as indicated in B were normalized by tubulin, and the level of AGO2 in shScramble mice was defined as 100%. Student's t-test was used. ∗, P < 0.05. D The levels of EV-A71 genomic RNAs in shScramble and shSTUB1 mice were detected by qRT-PCR. Student's t-test was used. ∗∗, P < 0.01. E–H Size distribution and abundance (counts per million of total vsRNA reads, CPM) of total vsRNAs sequenced from shScramble and shSTUB1 mice infected with EV-A71. Red, positive-stranded vsRNAs; blue, negative-stranded vsRNAs. The distribution of 22±1-nt vsRNA reads mapped to the (+)- and (−)-stranded EV-A71 genome and the relative abundances of (+)- and (−)-stranded vsRNAs are indicated. I–K The mRNA levels of IFN-α, IFN-β and INF-γ in shScramble and shSTUB1 mice were detected by qRT-PCR. Student's t-test was used. ns, no significant.
The effects of STUB1 knockdown on the replication of EV-A71 were examined by using the total RNAs extracted from the hind leg muscles of EV-A71-infected shSTUB1 and shScramble mice at 4 d.p.i. Our data showed that the viral RNA levels of EV-A71 were remarkably reduced in shSTUB1 mice compared with that in shScramble mice (Fig. 8D). To examine the status of antiviral RNAi, we examined the production of vsiRNAs in the same samples by using small RNA sequencing. As expected, the viral small RNA (vsRNA) reads from EV-A71-infected shScramble mice clustered as the Dicer cleavage products with an apparent peak in 22±1-nt size of both polarities and were enriched at the 5′-terminal region of EV-A71 genome (Fig. 8E and F and Table 1), confirming that EV-A71 infection elicits the production of vsiRNAs in newborn mice. Although vsRNA reads in EV-A71-infected shSTUB1 mice were less abundant than those in shScramble mice (Table 1), due to the reduced viral RNA replication, the relative abundance of 22±1-nt-size vsRNA reads of both polarities, especially the 22-nt-size, was remarkably increased in EV-A71-infected shSTUB1 mice (Fig. 8G and H and Table 1). These results showed that STUB1 deficiency enhanced the production of vsiRNAs and strengthened the antiviral RNAi in newborn mice infected with EV-A71. Besides, we showed that STUB1 deficiency had no or little effect on the induction of IFN-α, IFN-β and IFN-γ in the hind leg muscles of newborn mice after EV-A71 infection (Fig. 8I–K), consistent with the data obtained in shSTUB1 cells (Fig. 7) and excluded the potential interference of IFN-I system.
Table 1.
Profiles of viral small RNAs.
| Total Reads | miRNA | Total virus reads | (+)21-23 nt in total virus reads | % of (+)21-23 nt in total virus reads | (−)21-23 nt in total virus reads | % of (−)21-23 nt in total virus reads | |
|---|---|---|---|---|---|---|---|
| EV-A71 in shScramble mouse | 8377004 | 1812901 | 12253 | 2388 | 19.5 | 1193 | 9.74 |
| EV-A71 in shSTUB1 mouse | 16159735 | 5859088 | 6072 | 1482 | 24.4 | 872 | 14.4 |
Together, our findings indicate that STUB1 deficiency enhances antiviral RNAi, thereby inhibiting EV-A71 infection in newborn mice.
4. Discussion
RNAi is an evolutionarily conserved innate antiviral immune response from plants and invertebrates to mammals, while the knowledge about the regulation of antiviral RNAi in mammals remains to be elucidated. In this study, we demonstrated that the E3 ubiquitin ligase STUB1 was a new regulator to control the protein abundance of multiple components in the RNAi machinery, including Dicer, aviDicer and AGO1–4. Importantly, we provided the evidence that STUB1 deficiency enhanced the RNAi response during viral infection, which efficiently inhibited viral replication in an IFN-I independent manner in cells and in vivo. Our findings provided novel insights into the regulatory mechanism of antiviral RNAi in mammals.
Accumulative evidences have emerged in support that RNAi-dependent antiviral immunity is functional in mammals (Han et al., 2020b; Li et al., 2013, 2016; Qian et al., 2020; Qiu et al., 2017, 2020; Zhang et al., 2020b), and this extends a theme to demonstrate the regulatory mechanisms of antiviral RNAi. In support of it, we reported here that STUB1-mediated ubiquitination and proteasomal degradation of Dicer and AGO2 contributed to regulating the RNAi response during viral infection. In STUB1-deficient cells, both the shRNA- and siRNA-induced RNAi response were enhanced along with the increased protein levels of Dicer and AGO2, indicated that the up-regulation of Dicer and AGO2 mediated by STUB1 deficiency triggers the potency of RNAi. And this enhancing effect on the RNAi response showed potent antiviral activity in cells and in vivo. We proposed that the up-regulation of the pools of Dicer and AGO2 might ensure a timely response to dsRNA or siRNA by allowing large amounts of dsRNA to be cleaved by Dicer into siRNA or siRNA to be loaded into AGO2 to form RISC quickly. This speculation was supported by our data that the production of EV-A71 vsiRNAs was more abundant in shSTUB1 mice than that in shScramble mice, and the previous observations that siRNA production can be improved by increasing the protein level of Dicer in human and mouse cells (Demeter et al., 2019). Moreover, AGO4 has been recently identified to possess antiviral activity in mammals (Adiliaghdam et al., 2020). And aviDicer that triggers potent antiviral RNAi may express in pluripotent or multipotent stem cells in somatic tissues (Poirier et al., 2021). Considering that STUB1 can also target these two proteins, it may play important roles in regulating AGO4- and aviDicer-mediated antiviral effects as well. Therefore, our findings demonstrated that STUB1 controlled the potency of antiviral RNAi response by regulating the protein levels of the RNAi machinery in mammals.
In addition to ubiquitination and degradation, antiviral RNAi is also regulated by other mechanisms. Recent studies have suggested that antiviral RNAi may be negatively regulated by IFN-I response in mammalian somatic cells (Backes et al., 2014; Maillard et al., 2016). Our previous study also has uncovered that infection with ZIKV could induce the production of abundant vsiRNAs in hNPCs, which lack IFN-I response, but not in neurons differentiated from hNPC (Xu et al., 2019). Moreover, the RIG-I-like receptor LGP2 has been identified to block the long dsRNA processing of human and mouse Dicer in cells and in vivo (van der Veen et al., 2018; Zhang et al., 2021). Besides, the N-terminal helicase domain of human and mouse Dicer can auto inhibit its efficiency of dsRNA processing into siRNA (Flemr et al., 2013; Kennedy et al., 2015). Consistently, mouse and human stem cells have been recently found to encode aviDicer that lacks the N-termianl iHel domain to trigger abundant vsiRNA production and efficient antiviral RNAi (Poirier et al., 2021). Here, this work discovered that posttranslational modification of Dicer and AGO2 might serve as a novel mechanism for regulating the RNAi response during viral infection. Our findings, combined with others, demonstrated that antiviral RNAi was tightly controlled through the coupling of multiple forms of regulation in mammals.
The UPS plays important roles in the homeostatic control over the RNAi machinery in mammals. To date, a mere handful of E3 ubiquitin ligases have been identified to mediate Dicer and/or AGO degradation though UPS. It has been shown that Trim71 functions as an E3 ubiquitin ligase that facilitate proteasomal degradation of AGO2 in mouse stem cells (Rybak et al., 2009). Nevertheless, Trim71 is a stem cell factor that is not expressed in somatic cells, and its relevance to AGO2 ubiquitination and stability remains under debate (Chang et al., 2012; Chen et al., 2012). Moreover, recent studies have shown that the E3 ubiquitin ligase ZSWIM8 can induce AGO2 proteasomal degradation, but it recognizes and ubiquitinates the TDMD-associated-conformation of AGO2 (Han et al., 2020a; Shi et al., 2020). In the current study, we identified that the E3 ubiquitin ligase STUB1 served as a new regulator for the RNAi machinery, targeting Dicer, aviDicer and AGO1-4. Our data also showed that STUB1 degraded AGO2 irrespectively of ZSWIM8. Considering that STUB1 has been identified to ubiquitinate diverse substrates and ubiquitously express in different tissues or cell types (Zhang et al., 2020a), our findings suggested that STUB1-mediated degradation might be a general regulatory mechanism for controlling the protein abundance of the components in the RNAi machinery in mammals. Moreover, the observations that the expression level of endogenous STUB1 was stable in RD cells infected with EV-A71D23A suggested that the regulation of the RNAi machinery by STUB1 was a steady and constant regulatory pattern, highlighting the significant role of STUB1 in regulating the RNAi pathway in mammals.
5. Conclusions
In conclusion, our findings discovered that the E3 ubiquitin ligase, STUB1, was a new regulator for the RNAi machinery. Moreover, we provided the demonstration that STUB1-mediated degradation regulated the RNAi immune response during viral infection, providing novel insights into the regulation of antiviral RNAi in mammals.
Data availability
The RNA-seq data have been deposited to the NCBI GEO database under the accession number GSE189203.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Shumin Zhang: investigation, data curation, writing-original draft. Xuhua Zhang: investigation, methodology. Yuanyuan Bie: investigation, formal analysis. Jing Kong: investigation. An Wang: investigation. Yang Qiu: conceptualization, funding acquisition, writing-review&editing, supervision. Xi Zhou: conceptualization, funding acquisition, writing-review&editing, supervision.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgement
We sincerely thank all members of Zhou lab for their support.
This work was supported by the National Natural Science Foundation of China (31970169 to X.Z. and 82172269 and 81873964 to Y.Q.), the International Partnership Program of Chinese Academy of Sciences (153B42KYSB20200004 to X.Z.), the Young Top-notch Talent Cultivation Program of Hubei Province (Y.Q.), the Grant from the CAS Youth Innovation Promotion Association (2020332 to Y.Q.), the Hubei Province Natural Science Funds for Distinguished Young Scholar (2021CFA047 to Y.Q.), and the Young Top-notch Talent Cultivation Program of Hubei Province (Y.Q.).
Contributor Information
Yang Qiu, Email: yangqiu@wh.iov.cn.
Xi Zhou, Email: zhouxi@wh.iov.cn.
References
- Adiliaghdam F., Basavappa M., Saunders T.L., Harjanto D., Prior J.T., Cronkite D.A., Papavasiliou N., Jeffrey K.L. A requirement for Argonaute 4 in mammalian antiviral defense. Cell Rep. 2020;30:1690–1701. doi: 10.1016/j.celrep.2020.01.021. e1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S., Langlois R.A., Schmid S., Varble A., Shim J.V., Sachs D., tenOever B.R. The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014;8:114–125. doi: 10.1016/j.celrep.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y., Villarino A.V., Eisley C.J., Barbeau R., Barczak A.J., Heinz G.A., Kremmer E., Heissmeyer V., McManus M.T., Erle D.J., Rao A., Ansel K.M. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.M., Martinez N.J., Thornton J.E., Hagan J.P., Nguyen K.D., Gregory R.I. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lai F., Niswander L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 2012;26:803–815. doi: 10.1101/gad.187641.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter T., Vaskovicova M., Malik R., Horvat F., Pasulka J., Svobodova E., Flemr M., Svoboda P. Main constraints for RNAi induced by expressed long dsRNA in mouse cells. Life Sci Alliance. 2019;2 doi: 10.26508/lsa.201800289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Liu Z., Qiu Y., Kong J., Fu Y., Liu Y., Wang C., Quan J., Wang Q., Xu W., Yin L., Cui J., Xu Y., Curry S., Jiang S., Lu L., Zhou X. Inhibition of viral suppressor of RNAi proteins by designer peptides protects from enteroviral infection in vivo. Immunity. 2021;54:2231–2244. doi: 10.1016/j.immuni.2021.08.027. e2236. [DOI] [PubMed] [Google Scholar]
- Flemr M., Malik R., Franke V., Nejepinska J., Sedlacek R., Vlahovicek K., Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Li Y., Ding S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019;19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- Han J., LaVigne C.A., Jones B.T., Zhang H., Gillett F., Mendell J.T. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. 2020;370 doi: 10.1126/science.abc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Chen G., Wang J., Jee D., Li W.X., Lai E.C., Ding S.W. Mechanism and function of antiviral RNA interference in mice. mBio. 2020;11:e03278-19. doi: 10.1128/mBio.03278-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.M., Whisnant A.W., Kornepati A.V., Marshall J.B., Bogerd H.P., Cullen B.R. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E6945–E6954. doi: 10.1073/pnas.1513421112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H.H., Lin L.J., Hung L.Y., Chen P.S. Role of Dicer in regulating oxaliplatin resistance of colon cancer cells. Biochem. Biophys. Res. Commun. 2018;506:87–93. doi: 10.1016/j.bbrc.2018.10.071. [DOI] [PubMed] [Google Scholar]
- Li Y., Basavappa M., Lu J., Dong S., Cronkite D.A., Prior J.T., Reinecker H.C., Hertzog P., Han Y., Li W.X., Cheloufi S., Karginov F.V., Ding S.W., Jeffrey K.L. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol. 2016;2:16250. doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu J., Han Y., Fan X., Ding S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chen X., Novak M.K., Zhang S., Hu W. Repressing Ago2 mRNA translation by Trim71 maintains pluripotency through inhibiting let-7 microRNAs. Elife. 2021;10 doi: 10.7554/eLife.66288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W., Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P.V., Van der Veen A.G., Deddouche-Grass S., Rogers N.C., Merits A., Reis E Sousa C. Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J. 2016;35:2505–2518. doi: 10.15252/embj.201695086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier E.Z., Buck M.D., Chakravarty P., Carvalho J., Frederico B., Cardoso A., Healy L., Ulferts R., Beale R., Reis E.S.C. An isoform of Dicer protects mammalian stem cells against multiple RNA viruses. Science. 2021;373:231–236. doi: 10.1126/science.abg2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q., Zhou H., Shu T., Mu J., Fang Y., Xu J., Li T., Kong J., Qiu Y., Zhou X. The capsid protein of semliki forest virus antagonizes RNA interference in mammalian cells. J. Virol. 2020;94 doi: 10.1128/JVI.01233-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Xu Y., Zhang Y., Zhou H., Deng Y.Q., Li X.F., Miao M., Zhang Q., Zhong B., Hu Y., Zhang F.C., Wu L., Qin C.F., Zhou X. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity. 2017;46:992–1004. doi: 10.1016/j.immuni.2017.05.006. e1005. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Xu Y.P., Wang M., Miao M., Zhou H., Xu J., Kong J., Zheng D., Li R.T., Zhang R.R., Guo Y., Li X.F., Cui J., Qin C.F., Zhou X. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E.A., Michel G., Nitsch R., Krappmann D., Wulczyn F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Sanchez-David R.Y., Maillard P.V. Unlocking the therapeutic potential of antiviral RNAi. Immunity. 2021;54:2180–2182. doi: 10.1016/j.immuni.2021.09.013. [DOI] [PubMed] [Google Scholar]
- Shahrudin S., Ding S.W. Boosting stem cell immunity to viruses. Science. 2021;373:160–161. doi: 10.1126/science.abj5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.Y., Kingston E.R., Kleaveland B., Lin D.H., Stubna M.W., Bartel D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science. 2020;370 doi: 10.1126/science.abc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Yang J.S., Azzam G., Liu J.L., Lai E.C. Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 2013;20:789–795. doi: 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Zhang S., Qin X., Chang X., Cui X., Li H., Zhang S., Gao H., Wang P., Zhang Z., Luo J., Li Z. Foot-and-mouth disease virus capsid protein VP2 activates the cellular EIF2S1-ATF4 pathway and induces autophagy via HSPB1. Autophagy. 2018;14:336–346. doi: 10.1080/15548627.2017.1405187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah K., Chen S., Lu J., Wang X., Liu Q., Zhang Y., Long Y., Hu Z., Xu G. The E3 ubiquitin ligase STUB1 attenuates cell senescence by promoting the ubiquitination and degradation of the core circadian regulator BMAL1. J. Biol. Chem. 2020;295:4696–4708. doi: 10.1074/jbc.RA119.011280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen A.G., Maillard P.V., Schmidt J.M., Lee S.A., Deddouche-Grass S., Borg A., Kjaer S., Snijders A.P., Reis e Sousa C. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018;37 doi: 10.15252/embj.201797479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Kong J., Lyu B., Wang X., Qian Q., Zhou X., Qiu Y. The capsid protein of rubella virus antagonizes RNA interference in mammalian cells. Viruses. 2021;13:154. doi: 10.3390/v13020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.P., Qiu Y., Zhang B., Chen G., Chen Q., Wang M., Mo F., Xu J., Wu J., Zhang R.R., Cheng M.L., Zhang N.N., Lyu B., Zhu W.L., Wu M.H., Ye Q., Zhang D., Man J.H., Li X.F., Cui J., Xu Z., Hu B., Zhou X., Qin C.F. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019;29:265–273. doi: 10.1038/s41422-019-0152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa T., Takahashi H., Shikata S., Liu X., Tamura M., Asada S., Fukushima T., Fukuyama T., Tanaka Y., Sawasaki T., Kitamura T., Goyama S. The ubiquitin ligase STUB1 regulates stability and activity of RUNX1 and RUNX1-RUNX1T1. J. Biol. Chem. 2017;292:12528–12541. doi: 10.1074/jbc.M117.785675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Dong S., Luo Z., Xie X., Fu B., Li P., Liu C., Yang X., Chen Y., Wang X., Liu Z., Wu J., Yan Y., Wang F., Chen J.F., Zhang J., Long G., Goldman S.A., Li S., Zhao Z., Liang Q. The Zika virus capsid disrupts corticogenesis by suppressing dicer activity and miRNA biogenesis. Cell Stem Cell. 2020;27:618–632. doi: 10.1016/j.stem.2020.07.012. e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Hu Z.W., Mao C.Y., Shi C.H., Xu Y.M. CHIP as a therapeutic target for neurological diseases. Cell Death Dis. 2020;11:727. doi: 10.1038/s41419-020-02953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li Z., Ye Z., Xu Y., Wang B., Wang C., Dai Y., Lu J., Lu B., Zhang W., Li Y. The activation of antiviral RNA interference not only exists in neural progenitor cells but also in somatic cells in mammals. Emerg. Microb. Infect. 2020;9:1580–1589. doi: 10.1080/22221751.2020.1787798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu Y., Dai Y., Li Z., Wang J., Ye Z., Ren Y., Wang H., Li W.X., Lu J., Ding S.W., Li Y. Efficient Dicer processing of virus-derived double-stranded RNAs and its modulation by RIG-I-like receptor LGP2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data have been deposited to the NCBI GEO database under the accession number GSE189203.