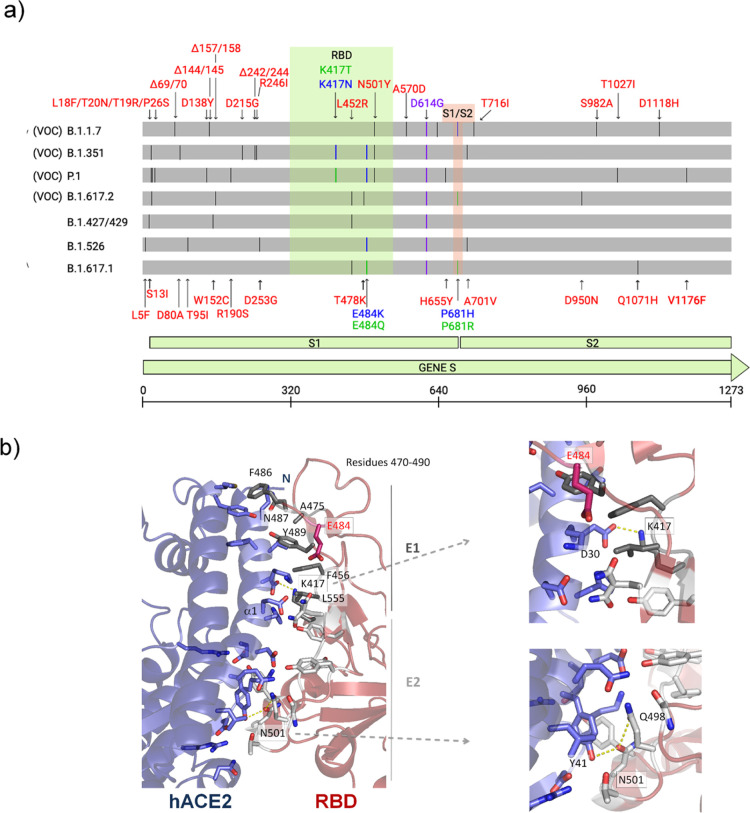
Figure 1.
Illustration of mutations in the S protein gene in different variants of SARS-CoV-2. (a) The RBD (residues 333–526) and the furin cleavage site (S1/S2) (residues 682–685) are colored in light green and salmon boxes, respectively. All mutations are represented by black lines and typed in red. Mutations that are found to be mutated for different residues are colored in blue and green. The green line shows different mutations in the same location. All SARS-CoV-2 variants shown in the figure have D614G, an advantage mutation for SARS-CoV-2. D614G has enhanced spike stability and transmission but does not significantly increase the binding affinity for hACE2 at 37 °C.37−39 VOC: variant of concern. (b) Cartoon representation of the RBD/hACE2 interface complex (PDB ID: 6M0J4). RDB presents two different regions, E1 (residues 417, 455–456, and 470–490) and E2 (444–454 and 493–505). The residues mutated in RBD of SARS-CoV-2 variants are K417, E484 (does not make part of the interface), and N501. RBDB.1.1.7 (alpha variant) has the mutation N501Y; RBDB.1.351 (beta variant) has K417N, E484K, and N501Y mutations; and RBDP.1 (gamma variant) has K417T, E484K, and N501Y mutations.