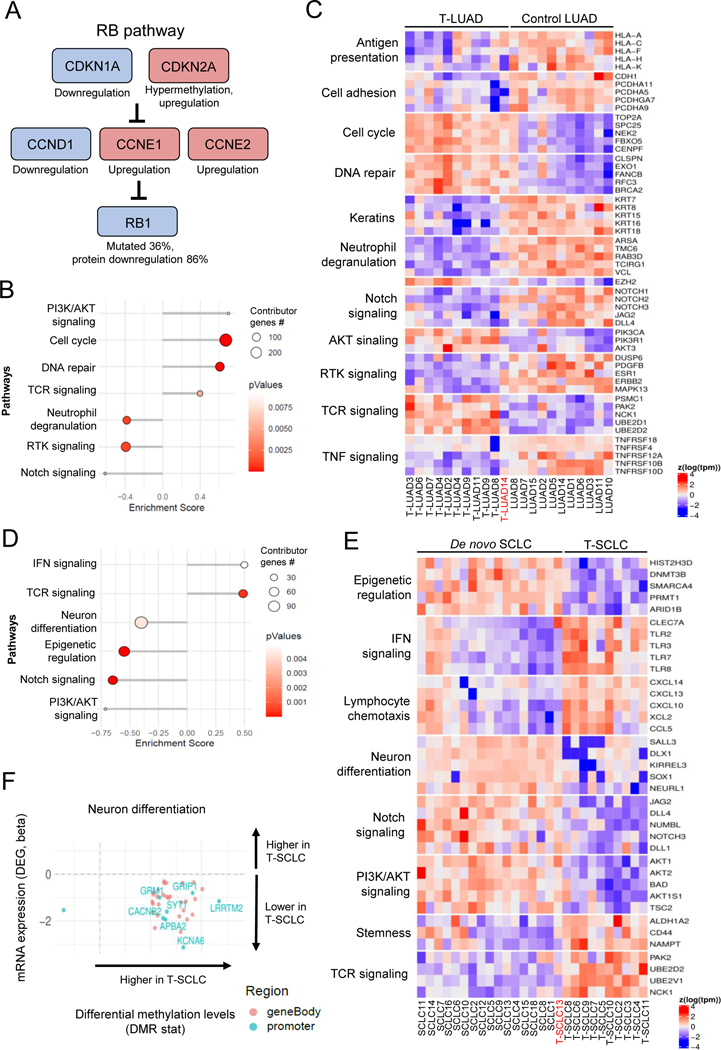
Figure 6. Integrative RNA and methylation analyses of T-LUAD and T-LUSC versus their control counterparts.
Related to Supplementary Figure S6 (A) Alterations in the RB pathway identified in T-LUAD. (B) Pathway enrichment analyses on the DEGs of the T-LUAD versus control LUAD comparison. (C) Heatmap highlighting DEGs of interest, grouped by recurrent pathways, of the T-LUAD versus control LUAD comparison. (D) Pathway enrichment analyses on the DEGs of T-SCLC versus de novo SCLC comparison. (E) Heatmap highlighting DEGs of interest, grouped by recurrent pathways, of T-SCLC versus de novo SCLC comparison. (F) Scatter plots showing DEGs exhibiting differential methylation levels in T-SCLC versus de novo SCLC comparison, grouped by pathways of interest. Significantly differentially expressed (q value < 0.05 and beta >= log2(1.5)) and methylated (FDR < 0.5 and differential methylation level greater than 0.1) sites are highlighted. Those genes where increased gene body or promoter methylation is correlated to expression positively and negatively, respectively, are labeled. Samples IDs in black and red indicate that they come from a combined histology specimen or a pre-/post-transformation specimen, respectively.