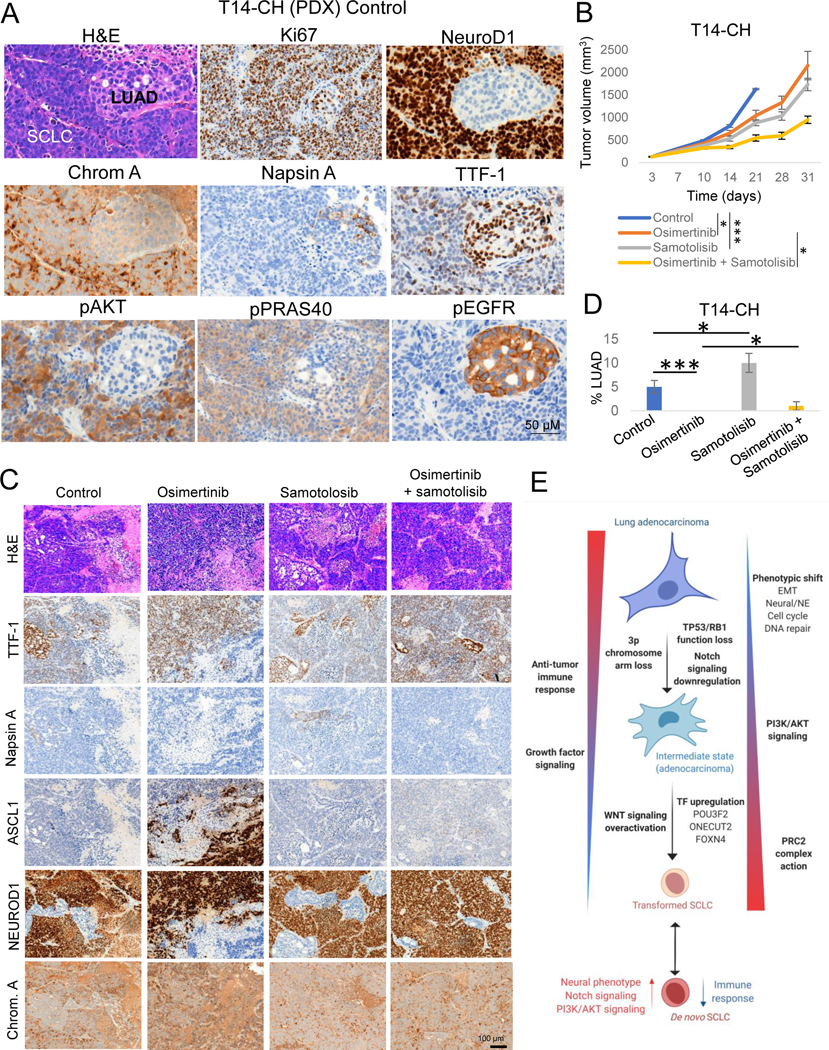
Figure 7. Potential therapeutic approaches for SCLC transformation.
Related to Supplementary Figure S7. (A) H&E and IHC markers of interest images showing combined LUAD and SCLC histology in the T14-CH PDX. (B) In vivo tumor growth of the combined LUAD/NE EGFR-mutant PDX model T14-CH with the EGFR inhibitor Osimertinib, the AKT inhibitor Samotolisib, or their combination. Group mean tumor size ± SEM is shown. Statistical differences in tumor sizes were assessed by a two-tailed Studentś t-test, using the tumor sizes for day 21 (control group endpoint) for those comparisons involving the control group, and on day 31 (experiment endpoint) for those comparisons involving the Osimertinib-treated group. (C) Representative H&E and IHC stains for the LUAD markers TTF-1 and Napsin A and the NE markers ASCL1, NEUROD1 and Chromogranin A, of tumors in each treatment arm. (D) Percentages of LUAD component per treatment group, showing the median ± standard deviation per group. Statistical differences were assessed by a two-tailed Studentś t-test. Diagnosis of each histological component was performed by a pathologist using morphological criteria and differential staining of LUAD (TTF-1, Napsin A), NE (ASCL1, NEUROD1, Chromogranin A) and other supporting (Ki67, pEGFR) markers. (E) Schematic of molecular and phenotype changes on the different steps of SCLC transformation. Our data suggest that transformation from LUAD to SCLC may be a progressive process involving multiple signaling pathways and phenotypic changes. This process may be initiated by the loss of TP53 and RB1, decreased dependence on RTK signaling and Notch signaling downregulation, and involve progressive activation of AKT and WNT signaling pathways, epigenomic regulation by the PRC2 complex and a number of additional epigenetic enzymes, acquisition of a neuronal and EMT phenotype, and downregulation of genes involved in multiple immune response pathways. Created with BioRender.com.p-values legend: * p<0.05, **p<0.01, ***<0.001.