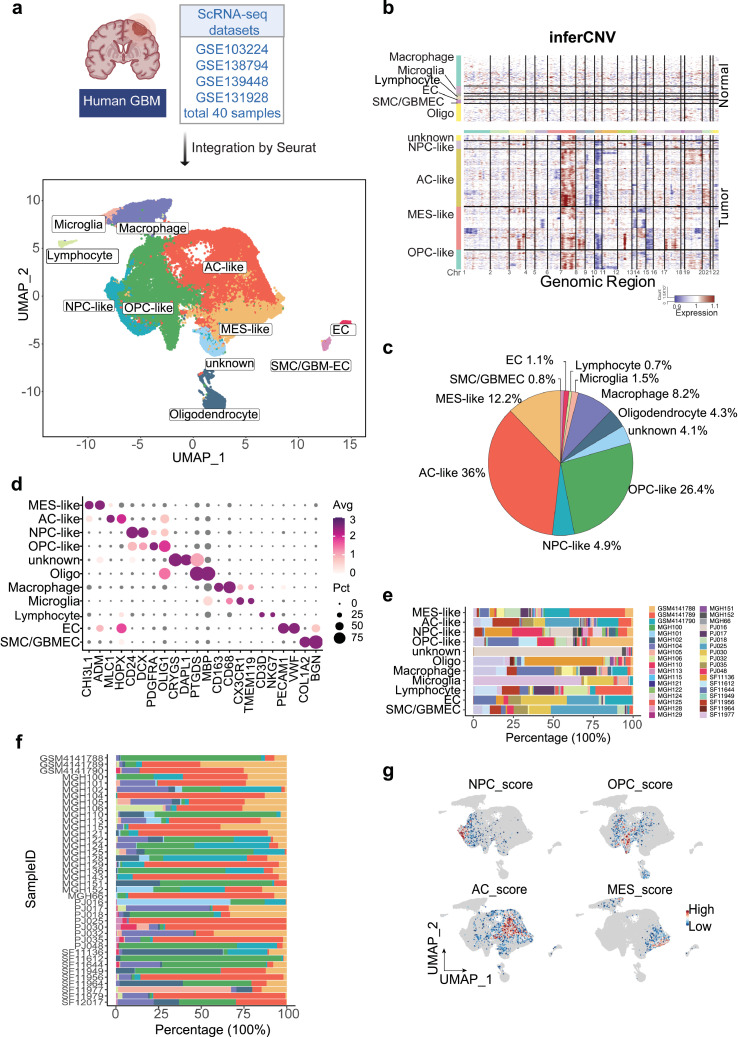
Figure 1.
Transcriptomic characterization of the TME and tumor heterogeneity in GBM by scRNA-seq analysis.
(a) Integration of 40 human GBM scRNA-seq data collected from four individual datasets by Seurat. A total of 54,764 cells were analyzed using UMAP, and 10 significant cell clusters are color-coded and labeled as indicated. (b) Inference of copy number variation analysis based on average expression of 100 genes shows the chromosome 7 gain (red) and chromosome 10 (blue) loss in tumor cells compared with normal cells. Each row corresponds to a cell type. (c) A pie chart demonstrates the proportions of each cell type in GBM. (d) Dot plot displays the represented markers for each major cell cluster, including MES-like, AC-like, NPC-like, OPC-like, oligo, macrophages, microglia, lymphocyte, EC, and GBM-ECs. (e) Stacked bar plots show the percentage of each cell cluster by patient ID and (f) the distribution of cell population in each patient. Cell type colors match those represented in the UMAP plot, while each patient is represented by a distinct color as indicated. (g) Feature plot displays the heterogeneity of GBM subtypes by Z-score, as determined by calculating the geometric mean of gene signature expressions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)