Figure 3.
Comparison of the 5-fold cross-validation predictions of a model trained with and without gene expression values
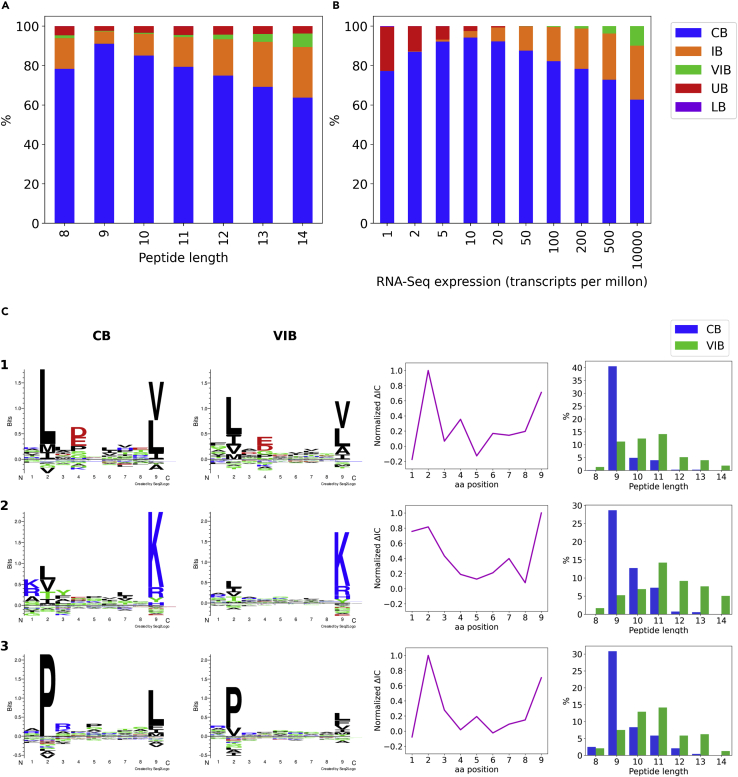
MS ligands from datasets A-D were classified according to the predictions obtained by model MS(woexp):HPA+MS(wexp):INT and its counterpart trained without gene expression MS(woexp+wexp).
(A) shows the peptide length distribution of the defined MS ligand subsets and (B) depicts the gene expression values distribution for the same groups. Finally, 3 exemplary alleles were chosen to illustrate the binding preferences of conserved binders (CB) and very improved binders (VIB).
(C) Characteristics of two groups of ligands: 1. their corresponding sequence logos (left), 2. the difference in information content for each position between the two logos (ΔIC = ICCB – ICVIB) normalized by maximum ΔIC across all positions (centre), and 3. their peptide length distributions (right). Exemplary alleles: 1. HLA-A∗02:01 (n = 379), 2. HLA-A∗03:01 (n = 267) and 3. HLA-B∗07:02 (n = 120). The logos for both CB and VIB binders are constructed with the same number of MS ligands, as specified before. CB, conserved binder; IB, improved binder; VIB, very improved binder; UB, unimproved binder; LB, lost binder.